4NBB
 
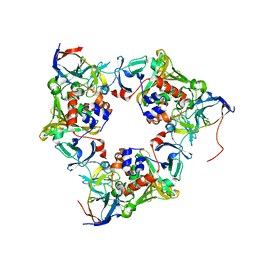 | | Carbazole- and oxygen-bound oxygenase with Ile262 replaced by Val and ferredoxin complex of carbazole 1,9a-dioxygenase | | Descriptor: | 9H-CARBAZOLE, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBE
 
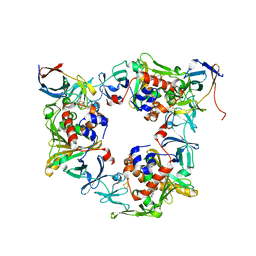 | | Fluorene-bound oxygenase with Phe275 replaced by Trp and ferredoxin complex of carbazole 1,9a-dioxygenase (form2) | | Descriptor: | 9H-fluorene, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
5AVF
 
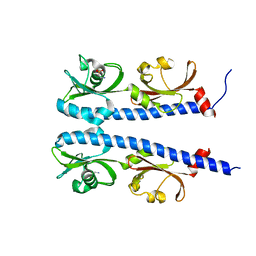 | | The ligand binding domain of Mlp37 with taurine | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Methyl-accepting chemotaxis (MCP) signaling domain protein | | Authors: | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
4NB8
 
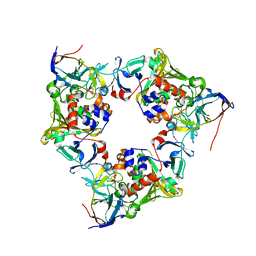 | | Oxygenase with Ile262 replaced by Leu and ferredoxin complex of carbazole 1,9a-dioxygenase | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin CarAc, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBA
 
 | | Carbazole-bound oxygenase with Ile262 replaced by Val and ferredoxin complex of carbazole 1,9a-dioxygenase | | Descriptor: | 9H-CARBAZOLE, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ashikawa, Y, Usami, Y, Inoue, K, Nojiri, H. | | Deposit date: | 2013-10-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase.
Appl.Environ.Microbiol., 80, 2014
|
|
7XUD
 
 | | Structure of G9a in complex with compound 26a | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUA
 
 | | Structure of G9a in complex with compound 10a | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUC
 
 | | Structure of G9a in complex with compound 11a | | Descriptor: | 1,2-ETHANEDIOL, 3,6,6-trimethyl-4-oxidanylidene-~{N}-[(2~{S})-1-oxidanylidene-1-phenylazanyl-hexan-2-yl]-5,7-dihydro-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUB
 
 | | Structure of G9a in complex with compound 10d | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
8KI1
 
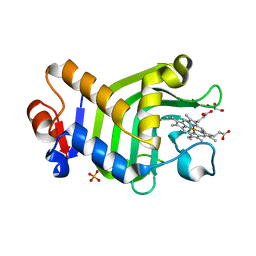 | | Crystal structure of the holo form of the hemophore HasA from Pseudomonas protegens Pf-5 | | Descriptor: | GLYCEROL, Heme acquisition protein HasAp, PHOSPHATE ION, ... | | Authors: | Shisaka, Y, Inaba, H, Sugimoto, H, Shoji, O. | | Deposit date: | 2023-08-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Heme-substituted protein assembly bridged by synthetic porphyrin: achieving controlled configuration while maintaining rotational freedom.
Rsc Adv, 14, 2024
|
|
8KI0
 
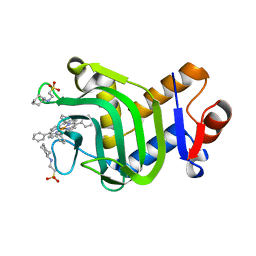 | | Crystal structure of the hemophore HasA from Pseudomonas protegens Pf-5 capturing Fe-tetraphenylporphyrin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Heme acquisition protein HasAp, ... | | Authors: | Shisaka, Y, Inaba, H, Sugimoto, H, Shoji, O. | | Deposit date: | 2023-08-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Heme-substituted protein assembly bridged by synthetic porphyrin: achieving controlled configuration while maintaining rotational freedom.
Rsc Adv, 14, 2024
|
|
5C5O
 
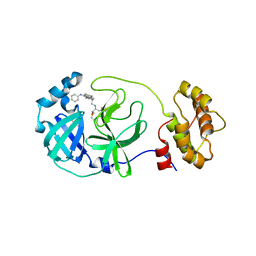 | | Structure of SARS-3CL protease complex with a phenyl-beta-alanyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-({[(3S,4aR,8aS)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2015-06-21 | | Release date: | 2016-06-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
to be published
|
|
8S5F
 
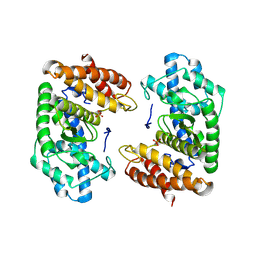 | | Crystal structure of the HExxH domain of ChlBHExxH a novel alpha-ketoglutarate dependent oxygenase | | Descriptor: | ChlH from Chlorogloeopsis sp., PHOSPHATE ION | | Authors: | de la Mora, E, Amara, P, Usclat, A, Morishita, Y, Morinaka, B, Nicolet, Y. | | Deposit date: | 2024-02-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Fused radical SAM and alpha KG-HExxH domain proteins contain a distinct structural fold and catalyse cyclophane formation and beta-hydroxylation.
Nat.Chem., 2024
|
|
5KC2
 
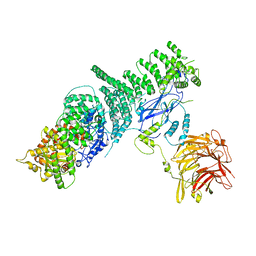 | | Negative stain structure of Vps15/Vps34 complex | | Descriptor: | Phosphatidylinositol 3-kinase VPS34, Serine/threonine-protein kinase VPS15 | | Authors: | Kirsten, M.L, Zhang, L, Ohashi, Y, Perisic, O, Williams, R.L, Sachse, C. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
5C5N
 
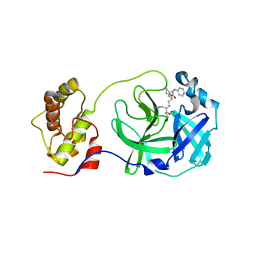 | | Structure of SARS-3CL protease complex with a phenyl-beta-alanyl (R,S)-N-decalin type inhibitor | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-({[(3R,4aS,8aR)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2015-06-21 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
to be published
|
|
6R9W
 
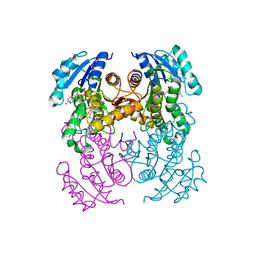 | | Crystal structure of InhA in complex with AP-124 inhibitor | | Descriptor: | (2~{S})-1-(benzimidazol-1-yl)-3-(2,3-dihydro-1~{H}-inden-5-yloxy)propan-2-ol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Takebayashi, Y, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-04-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of New and Potent InhA Inhibitors as Antituberculosis Agents: Structure-Based Virtual Screening Validated by Biological Assays and X-ray Crystallography.
J.Chem.Inf.Model., 60, 2020
|
|
6RUJ
 
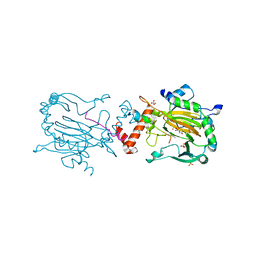 | |
5ETY
 
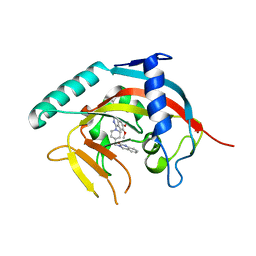 | | Crystal Structure of human Tankyrase-1 bound to K-756 | | Descriptor: | 3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-1,4-dihydroquinazolin-2-one, Tankyrase-1, ZINC ION | | Authors: | Takahashi, Y, Miyagi, H, Suzuki, M, Saito, J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery and Characterization of K-756, a Novel Wnt/ beta-Catenin Pathway Inhibitor Targeting Tankyrase
Mol.Cancer Ther., 15, 2016
|
|
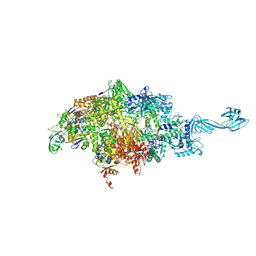
6WOX
 
 | |
6WOY
 
 | |
3D79
 
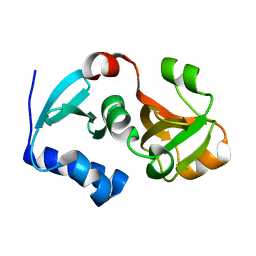 | | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3 | | Descriptor: | Putative uncharacterized protein PH0734 | | Authors: | Nishimura, Y, Miyazono, K, Sawano, Y, Makino, T, Nagata, K, Tanokura, M. | | Deposit date: | 2008-05-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of hypothetical protein PH0734.1 from hyperthermophilic archaea Pyrococcus horikoshii OT3.
Proteins, 73, 2008
|
|
5B3D
 
 | | Structure of a flagellar type III secretion chaperone, FlgN | | Descriptor: | Flagella synthesis protein FlgN | | Authors: | Nakanishi, Y, Kinoshita, M, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-02-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rearrangements of alpha-helical structures of FlgN chaperone control the binding affinity for its cognate substrates during flagellar type III export
Mol.Microbiol., 101, 2016
|
|
8K71
 
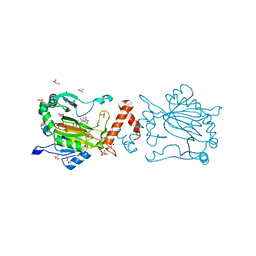 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((2-((naphthalen-2-ylmethyl)sulfonyl)acetyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | Descriptor: | 2-[(2~{Z})-2-[2-(naphthalen-2-ylmethylsulfonyl)ethanoylimino]-3-oxidanyl-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
8K72
 
 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((3-(phenylsulfonamido)propanoyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | Descriptor: | 2-[(2~{Z})-3-oxidanyl-2-[3-(phenylsulfonylamino)propanoylimino]-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
8K73
 
 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((1-(phenylsulfonyl)pyrrolidine-3-carbonyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | Descriptor: | 2-[(2~{Z})-3-oxidanyl-2-[(3~{R})-1-(phenylsulfonyl)pyrrolidin-3-yl]carbonylimino-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
