8X6B
 
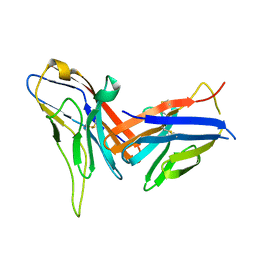 | | Crystal structure of immune receptor PVRIG in complex with ligand Nectin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nectin-2, Transmembrane protein PVRIG | | Authors: | Hu, S.T, Han, P, Wang, H, Qi, J.X. | | Deposit date: | 2023-11-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the immune recognition and selectivity of the immune receptor PVRIG for ligand Nectin-2.
Structure, 2024
|
|
7O4Z
 
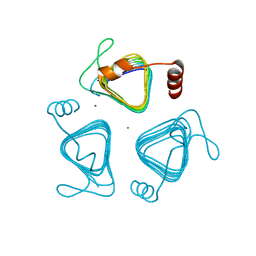 | | Crystal structure of the carbonic anhydrase-like domain of CcmM from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | CHLORIDE ION, Carboxysome assembly protein CcmM, NICKEL (II) ION | | Authors: | Zang, K, Wang, H, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2021-04-07 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Scaffolding protein CcmM directs multiprotein phase separation in beta-carboxysome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7O54
 
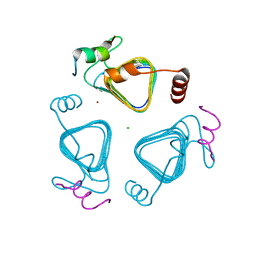 | | Crystal structure of the carbonic anhydrase-like domain of CcmM in complex with the C-terminal 17 residues of CcaA from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | CHLORIDE ION, Carbonic anhydrase, Carboxysome assembly protein CcmM, ... | | Authors: | Zang, K, Wang, H, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2021-04-07 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Scaffolding protein CcmM directs multiprotein phase separation in beta-carboxysome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3DD4
 
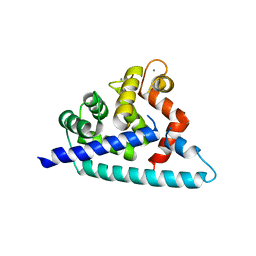 | |
6TDA
 
 | | Structure of SWI/SNF chromatin remodeler RSC bound to a nucleosome | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Wagner, F.R, Dienemann, C, Wang, H, Stuetzer, A, Tegunov, D, Urlaub, H, Cramer, P. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome.
Nature, 579, 2020
|
|
8JIJ
 
 | | Alanine decarboxylase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine decarboxylase, ZINC ION | | Authors: | Gong, W, Wang, H. | | Deposit date: | 2023-05-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of alanine decarboxylase
To Be Published
|
|
8JIK
 
 | | Alanine decarboxylase | | Descriptor: | CALCIUM ION, ETHANAMINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Gong, W, Wang, H. | | Deposit date: | 2023-05-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of alanine decarboxylase
To Be Published
|
|
1A8O
 
 | | HIV CAPSID C-TERMINAL DOMAIN | | Descriptor: | HIV CAPSID | | Authors: | Gamble, T.R, Yoo, S, Vajdos, F.F, Von Schwedler, U.K, Worthylake, D.K, Wang, H, Mccutcheon, J.P, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein.
Science, 278, 1997
|
|
1A43
 
 | | STRUCTURE OF THE HIV-1 CAPSID PROTEIN DIMERIZATION DOMAIN AT 2.6A RESOLUTION | | Descriptor: | HIV-1 CAPSID | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-02-10 | | Release date: | 1999-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BAJ
 
 | | HIV-1 CAPSID PROTEIN C-TERMINAL FRAGMENT PLUS GAG P2 DOMAIN | | Descriptor: | GAG POLYPROTEIN | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1AUM
 
 | | HIV CAPSID C-TERMINAL DOMAIN (CAC146) | | Descriptor: | HIV CAPSID | | Authors: | Hill, C.P, Gamble, T.R, Yoo, S, Vajdos, F.F, Von Schwedler, U.K, Worthylake, D.K, Wang, H, Mccutcheon, J.P, Sundquist, W.I. | | Deposit date: | 1997-08-29 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein.
Science, 278, 1997
|
|
1BC9
 
 | | CYTOHESIN-1/B2-1 SEC7 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOHESIN-1 | | Authors: | Betz, S.F, Schnuchel, A, Wang, H, Olejniczak, E.T, Meadows, R.P, Fesik, S.W. | | Deposit date: | 1998-05-06 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cytohesin-1 (B2-1) Sec7 domain and its interaction with the GTPase ADP ribosylation factor 1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1Q2U
 
 | | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Huai, Q, Sun, Y, Wang, H, Chin, L.S, Li, L, Robinson, H, Ke, H. | | Deposit date: | 2003-07-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease
Febs Lett., 549, 2003
|
|
4QC2
 
 | | Crystal structure of lipopolysaccharide transport protein LptB in complex with ATP and Magnesium ions | | Descriptor: | ABC transporter related protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, Z, Xiang, Q, Zhu, X, Dong, H, He, C, Wang, H, Zhang, Y, Wang, W, Dong, C. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and functional studies of conserved nucleotide-binding protein LptB in lipopolysaccharide transport.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4AOJ
 
 | | Human TrkA in complex with the inhibitor AZ-23 | | Descriptor: | 5-chloranyl-N2-[(1S)-1-(5-fluoranylpyridin-2-yl)ethyl]-N4-(3-propan-2-yloxy-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR, ZINC ION | | Authors: | Wang, T, Lamb, M.L, Block, M.H, Davies, A.M, Han, Y, Hoffmann, E, Ioannidis, S, Josey, J.A, Liu, Z, Lyne, P.D, MacIntyre, T, Mohr, P.J, Omer, C.A, Sjogren, T, Thress, K, Wang, B, Wang, H, Yu, D, Zhang, H. | | Deposit date: | 2012-03-28 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Disubstituted Imidazo[4,5-B]Pyridines and Purines as Potent Trka Inhibitors
Acs Med.Chem.Lett., 3, 2012
|
|
5GTX
 
 | | Crystal structure of mutated buckwheat glutaredoxin | | Descriptor: | buckwheat glutaredoxin | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
3C4A
 
 | | Crystal structure of vioD hydroxylase in complex with FAD from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR158 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable tryptophan hydroxylase vioD | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Wang, H, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-05 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of vioD hydroxylase in complex with FAD from Chromobacterium violaceum.
To be Published
|
|
5GOU
 
 | | Structure of a 16-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Wang, W, Wang, H, Zhao, G. | | Deposit date: | 2016-07-29 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Conversion of the Native 24-mer Ferritin Nanocage into Its Non-Native 16-mer Analogue by Insertion of Extra Amino Acid Residues.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
3FOK
 
 | | Crystal Structure of Cgl0159 From Corynebacterium glutamicum (Brevibacterium flavum). Northeast Structural Genomics Target CgR115 | | Descriptor: | uncharacterized protein Cgl0159 | | Authors: | Seetharaman, J, Neely, H, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-30 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Cgl0159 From Corynebacterium glutamicum (Brevibacterium flavum). Northeast Structural Genomics Target CgR115
To be Published
|
|
3GGN
 
 | | Crystal structure of DR_A0006 from Deinococcus radiodurans. Northeast Structural Genomics Consortium Target DrR147D | | Descriptor: | Uncharacterized protein DR_A0006 | | Authors: | Seetharaman, J, Neely, H, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-28 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of DR_A0006 from Deinococcus radiodurans.
To be Published
|
|
3GGM
 
 | | Crystal Structure of BT9727_2919 from Bacillus thuringiensis subsp. Northeast Structural Genomics Target BuR228B | | Descriptor: | uncharacterized protein BT9727_2919 | | Authors: | Seetharaman, J, Neely, H, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-28 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of BT9727_2919 from Bacillus thuringiensis subsp. Northeast Structural Genomics Target BuR228B
To be Published
|
|
3BJC
 
 | | Crystal structure of the PDE5A catalytic domain in complex with a novel inhibitor | | Descriptor: | 5-ethoxy-4-(1-methyl-7-oxo-3-propyl-6,7-dihydro-1H-pyrazolo[4,3-d]pyrimidin-5-yl)thiophene-2-sulfonamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, G, Wang, H, Howard, R, Cai, J, Wan, Y, Ke, H. | | Deposit date: | 2007-12-03 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An insight into the pharmacophores of phosphodiesterase-5 inhibitors from synthetic and crystal structural studies
BIOCHEM.PHARM., 75, 2008
|
|
2A4G
 
 | | Hepatitis C Protease NS3-4A serine protease with Ketoamide Inhibitor SCH225724 Bound | | Descriptor: | ({1-[1-CARBAMOYL-PHENYL-METHYL)-CARBAMOYL]-METHYL}-AMINOOXALYL)-BUTYLCARBAMOYL)-3-METHYL-BUTYLCARBAMOYL)-CYCLOHEXYL-METHYL)-CARBAMIC ACID ISOBUTYL ESTER, NS3 protease/helicase, NS4a peptide, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chan, T.Y, Bennett, F, Bogen, S.L, Chen, K, Gu, H, Hong, L, Jao, E, Liu, Y.T, Lovey, R.G, Parekh, T, Pike, R.E, Pinto, P, Santhanam, B, Venkatraman, S, Vaccaro, H, Wang, H, Yang, X, Zhu, Z, Mckittrick, B, Saksena, A.K, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Ingram, R, Malcolm, B, Prongay, A.J, Yao, N, Marten, B, Madison, V, Kemp, S, Levy, O, Lim-Wilby, M, Tamura, S, Ganguly, A.K. | | Deposit date: | 2005-06-28 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hepatitis C virus NS3-4a serine protease inhibitors. SAR of P2' moiety with improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
7YPN
 
 | | Crystal structure of transaminase CC1012 mutant M9 complexed with PLP | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase family protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, L, Wang, H, Wei, D. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism-Guided Computational Design of omega-Transaminase by Reprograming of High-Energy-Barrier Steps.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7YPM
 
 | | Crystal structure of transaminase CC1012 complexed with PLP and L-alanine | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aspartate aminotransferase family protein, ... | | Authors: | Yang, L, Wang, H, Wei, D. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Mechanism-Guided Computational Design of omega-Transaminase by Reprograming of High-Energy-Barrier Steps.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
