7F9H
 
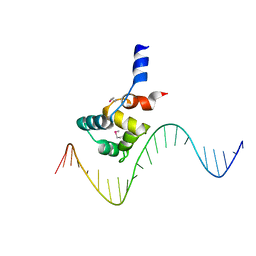 | | complex structure of EnrR-DNA | | Descriptor: | EnrR repressor, target DNA | | Authors: | Gan, J.H, Wang, Q.Y. | | Deposit date: | 2021-07-04 | | Release date: | 2022-05-11 | | Last modified: | 2022-05-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding.
Nucleic Acids Res., 50, 2022
|
|
6NPP
 
 | |
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
6NZR
 
 | |
6NZH
 
 | |
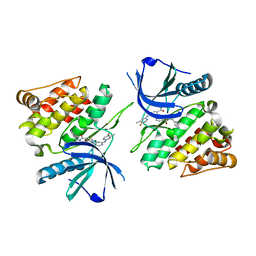
6NZQ
 
 | |
6NWZ
 
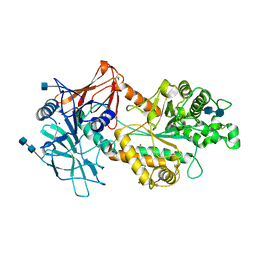 | | Crystal structure of Agd3 a novel carbohydrate deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Bamford, N.C, Howell, P.L. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical characterization of the exopolysaccharide deacetylase Agd3 required for Aspergillus fumigatus biofilm formation.
Nat Commun, 11, 2020
|
|
6NZF
 
 | |
7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
6NZE
 
 | |
6NZP
 
 | |
8J4T
 
 | | GajA-GajB complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Wei, T, Ma, J, Huo, Y. | | Deposit date: | 2023-04-20 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and biochemical insights into the mechanism of the Gabija bacterial immunity system.
Nat Commun, 15, 2024
|
|
6OJ1
 
 | | Crystal Structure of Aspergillus fumigatus Ega3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bamford, N.C, Subramanian, A.S, Millan, C, Uson, I, Howell, P.L. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
6OQ5
 
 | | Structure of the full-length Clostridium difficile toxin B in complex with 3 VHHs | | Descriptor: | 5D, 7F, E3, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7C2Q
 
 | | The crystal structure of COVID-19 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Hu, X.H, Zhou, H, Wang, Q.S, Li, j, Zhang, J. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of SARS-CoV-2 main protease in the apo state.
Sci China Life Sci, 64, 2021
|
|
7D0Q
 
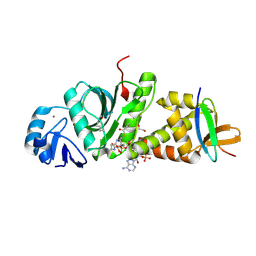 | |
7D0P
 
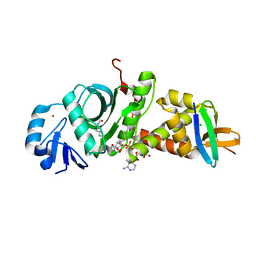 | |
7D0O
 
 | | Crystal structure of human HBO1-BRPF2 in apo form | | Descriptor: | 1,2-ETHANEDIOL, BRD1 protein, Histone acetyltransferase KAT7, ... | | Authors: | Li, W, Ding, J. | | Deposit date: | 2020-09-11 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | HBO1 is a versatile histone acyltransferase critical for promoter histone acylations.
Nucleic Acids Res., 49, 2021
|
|
7D0S
 
 | |
7D0R
 
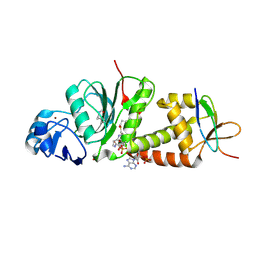 | |
7E0F
 
 | |
7E20
 
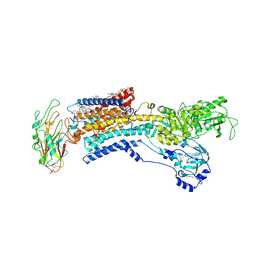 | | Cryo EM structure of a K+-bound Na+,K+-ATPase in the E2 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E21
 
 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state with ATP-gamma-S | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E1Z
 
 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7ENO
 
 | | Mutant strain M3 of foot-and-mouth disease virus type O | | Descriptor: | VP1 of O type FMDV capsid, VP2 of O type FMDV capsid, VP3 of O type FMDV capsid, ... | | Authors: | Dong, H, Lu, Y. | | Deposit date: | 2021-04-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | A Heat-Induced Mutation on VP1 of Foot-and-Mouth Disease Virus Serotype O Enhanced Capsid Stability and Immunogenicity.
J.Virol., 95, 2021
|
|
