7O82
 
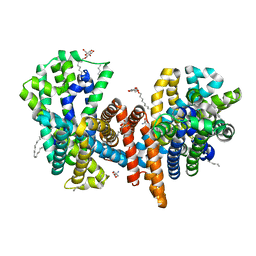 | | The L-arginine/agmatine antiporter from E. coli at 1.7 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine/agmatine antiporter, DECANE, ... | | Authors: | Jeckelmann, J.M, Ilgue, H, Kalbermatter, D, Fotiadis, D. | | Deposit date: | 2021-04-14 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution structure of the amino acid transporter AdiC reveals insights into the role of water molecules and networks in oligomerization and substrate binding.
Bmc Biol., 19, 2021
|
|
4AQY
 
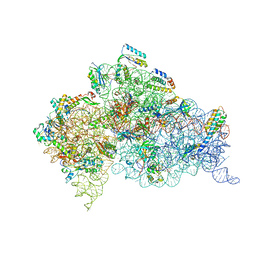 | | Structure of ribosome-apramycin complexes | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Matt, T, Ng, C.L, Lang, K, Sha, S.H, Akbergenov, R, Shcherbakov, D, Meyer, M, Duscha, S, Xie, J, Dubbaka, S.R, Perez-Fernandez, D, Vasella, A, Ramakrishnan, V, Schacht, J, Bottger, E.C. | | Deposit date: | 2012-04-20 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Dissociation of Antibacterial Activity and Aminoglycoside Ototoxicity in the 4-Monosubstituted 2-Deoxystreptamine Apramycin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5WIT
 
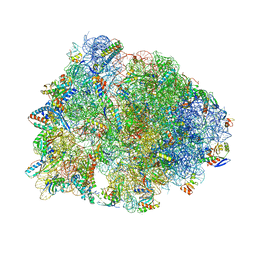 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pikromycin and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | (3R,5R,6S,7S,9R,11E,13S,14R)-14-ethyl-13-hydroxy-3,5,7,9,13-pentamethyl-2,4,10-trioxo-1-oxacyclotetradec-11-en-6-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Almutairi, M.M, Svetlov, M.S, Hansen, D.A, Khabibullina, N.F, Klepacki, D, Kang, H.Y, Sherman, D.H, Vazquez-Laslop, N, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2017-07-20 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins.
Nucleic Acids Res., 45, 2017
|
|
2XGL
 
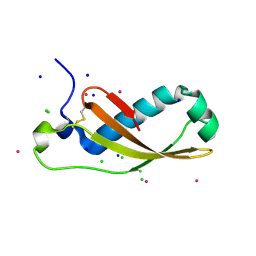 | | The X-ray structure of the Escherichia coli colicin M immunity protein demonstrates the presence of a disulphide bridge, which is functionally essential | | Descriptor: | CADMIUM ION, CHLORIDE ION, COLICIN-M IMMUNITY PROTEIN, ... | | Authors: | Gerard, F, Brooks, M.A, Barreteau, H, Touze, T, Graille, M, Bouhss, A, Blanot, D, Tilbeurgh, H.v, Mengin-Lecreulx, D. | | Deposit date: | 2010-06-07 | | Release date: | 2010-11-17 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-Ray Structure and Site-Directed Mutagenesis Analysis of the Escherichia Coli Colicin M Immunity Protein.
J.Bacteriol., 193, 2011
|
|
4A5O
 
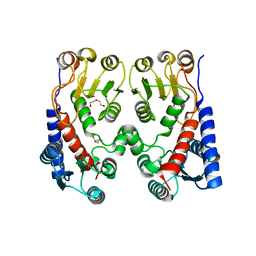 | | Crystal structure of Pseudomonas aeruginosa N5, N10- methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) | | Descriptor: | BIFUNCTIONAL PROTEIN FOLD, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Eadsforth, T.C, Gardiner, M, Maluf, F.V, McElroy, S, James, D, Frearson, J, Gray, D, Hunter, W.N. | | Deposit date: | 2011-10-26 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Assessment of Pseudomonas Aeruginosa N(5),N(10)-Methylenetetrahydrofolate Dehydrogenase - Cyclohydrolase as a Potential Antibacterial Drug Target.
Plos One, 7, 2012
|
|
2XHJ
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. SeMet form of vCBM60. | | Descriptor: | CALCIUM ION, CALCIUM-DEPENDENT CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
2XHH
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
6HPY
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 12 | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]propanoic acid, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
8ELC
 
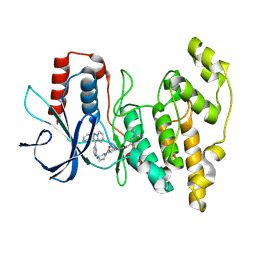 | | Human JNK2 bound to covalent inhibitor YL2056 | | Descriptor: | 4-(dimethylamino)-N-{4-[(3S)-3-({4-[(8R)-2-phenylpyrazolo[1,5-a]pyridin-3-yl]pyrimidin-2-yl}amino)pyrrolidine-1-carbonyl]phenyl}butanamide, Mitogen-activated protein kinase 9 | | Authors: | Li, L, Gurbani, D, Westover, K.D. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.072 Å) | | Cite: | Development of a Covalent Inhibitor of c-Jun N-Terminal Protein Kinase (JNK) 2/3 with Selectivity over JNK1.
J.Med.Chem., 66, 2023
|
|
6I0M
 
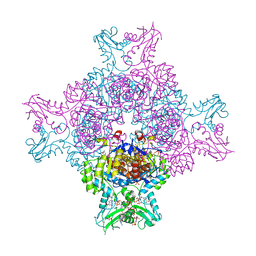 | | Structure of human IMP dehydrogenase, isoform 2, bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Buey, R.M, Fernandez-Justel, D, Revuelta, J.L. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.567 Å) | | Cite: | A Nucleotide-Dependent Conformational Switch Controls the Polymerization of Human IMP Dehydrogenases to Modulate their Catalytic Activity.
J. Mol. Biol., 431, 2019
|
|
6C98
 
 | | Crystal structure of FcRn bound to UCB-84 | | Descriptor: | 1-[7-(3-fluorophenyl)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-6-yl]ethan-1-one, Beta-2-microglobulin, CYSTEINE, ... | | Authors: | Fox III, D, Lukacs, C.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
1LNX
 
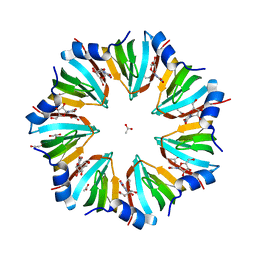 | | Crystal structure of the P.aerophilum SmAP1 heptamer in a new crystal form (C2221) | | Descriptor: | ACETIC ACID, GLYCEROL, URIDINE, ... | | Authors: | Mura, C, Kozhukhovsky, A, Eisenberg, D. | | Deposit date: | 2002-05-04 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs)
Protein Sci., 12, 2003
|
|
6TUA
 
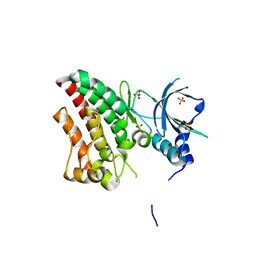 | | The RYK Pseudokinase Domain | | Descriptor: | SULFATE ION, Tyrosine-protein kinase RYK | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
4A29
 
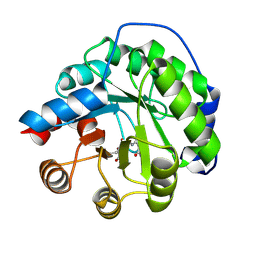 | | Structure of the engineered retro-aldolase RA95.0 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, D-MALATE, ENGINEERED RETRO-ALDOL ENZYME RA95.0 | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-23 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4A2R
 
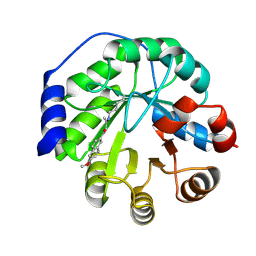 | | Structure of the engineered retro-aldolase RA95.5-5 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
6IAT
 
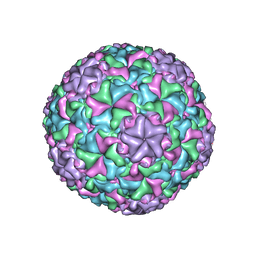 | | Icosahedrally averaged capsid of bacteriophage P68 | | Descriptor: | Arstotzka protein, Major head protein | | Authors: | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | Deposit date: | 2018-11-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
6ICE
 
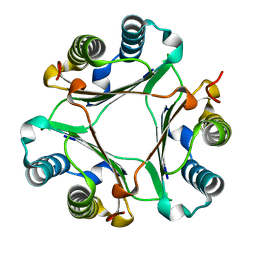 | | Crystal structure of Hamster MIF | | Descriptor: | Macrophage migration inhibitory factor | | Authors: | Sundaram, R, Vasudevan, D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macrophage migration inhibitory factor of Syrian golden hamster shares structural and functional similarity with human counterpart and promotes pancreatic cancer.
Sci Rep, 9, 2019
|
|
5FGP
 
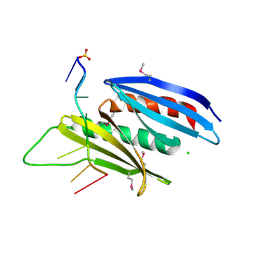 | | Crystal structure of D. melanogaster Pur-alpha repeat I-II in complex with DNA. | | Descriptor: | CG1507-PB, isoform B, CHLORIDE ION, ... | | Authors: | Weber, J, Janowski, R, Niessing, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of nucleic-acid recognition and double-strand unwinding by the essential neuronal protein Pur-alpha.
Elife, 5, 2016
|
|
2XRA
 
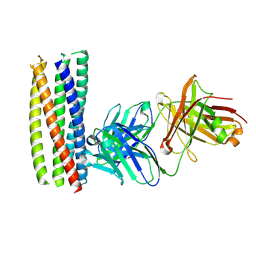 | | crystal structure of the HK20 Fab in complex with a gp41 mimetic 5- Helix | | Descriptor: | HK20, HUMAN MONOCLONAL ANTIBODY HEAVY CHAIN, HUMAN MONOCLONAL ANTIBODY LIGHT CHAIN, ... | | Authors: | Sabin, C, Corti, D, Buzon, V, Seaman, M.S, Lutje Hulsik, D, Hinz, A, Vanzetta, F, Agatic, G, Silacci, C, Langedijk, J.P.M, Mainetti, L, Scarlatti, G, Sallusto, F, Weiss, R, Lanzavecchia, A, Weissenhorn, W. | | Deposit date: | 2010-09-13 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and size-dependent neutralization properties of HK20, a human monoclonal antibody binding to the highly conserved heptad repeat 1 of gp41.
PLoS Pathog., 6, 2010
|
|
4AE7
 
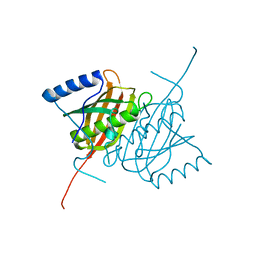 | | Crystal structure of human THEM5 | | Descriptor: | THIOESTERASE SUPERFAMILY MEMBER 5 | | Authors: | Zhuravleva, E, Gut, H, Hynx, D, Marcellin, D, Bleck, C.K.E, Genoud, C, Cron, P, Keusch, J.J, Dummler, B, Degli Esposti, M, Hemmings, B.A. | | Deposit date: | 2012-01-09 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Acyl Coenzyme a Thioesterase Them5/Acot15 is Involved in Cardiolipin Remodeling and Fatty Liver Development.
Mol.Cell.Biol., 32, 2012
|
|
4AE8
 
 | | Crystal structure of human THEM4 | | Descriptor: | THIOESTERASE SUPERFAMILY MEMBER 4 | | Authors: | Zhuravleva, E, Gut, H, Hynx, D, Marcellin, D, Bleck, C.K.E, Genoud, C, Cron, P, Keusch, J.J, Dummler, B, Degli Esposti, M, Hemmings, B.A. | | Deposit date: | 2012-01-09 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Acyl Coenzyme a Thioesterase Them5/Acot15 is Involved in Cardiolipin Remodeling and Fatty Liver Development.
Mol.Cell.Biol., 32, 2012
|
|
5NO4
 
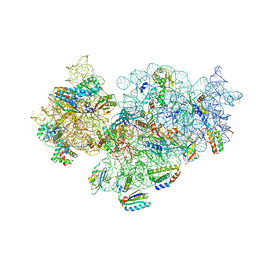 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate with uS3) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
5NO2
 
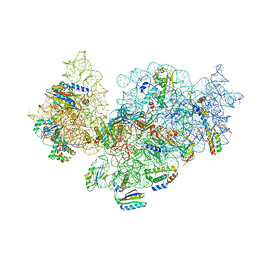 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
6C2Z
 
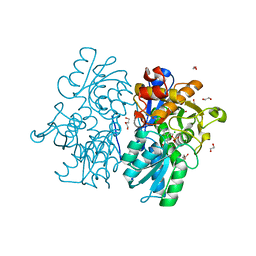 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-Aminoacrylate Intermediate | | Descriptor: | 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-09 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
7OK0
 
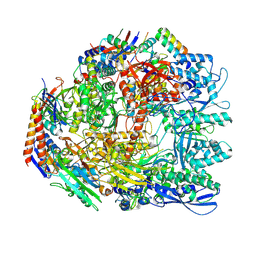 | | Cryo-EM structure of the Sulfolobus acidocaldarius RNA polymerase at 2.88 A | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-05-17 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
