3J5M
 
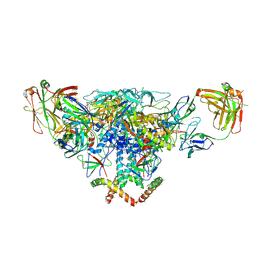 | | Cryo-EM structure of the BG505 SOSIP.664 HIV-1 Env trimer with 3 PGV04 Fabs | | Descriptor: | BG505 SOSIP gp120, BG505 SOSIP gp41, PGV04 heavy chain, ... | | Authors: | Lyumkis, D, Julien, J.-P, Wilson, I.A, Ward, A.B. | | Deposit date: | 2013-10-26 | | Release date: | 2013-11-13 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
3GMR
 
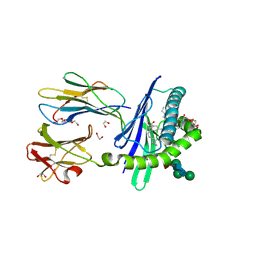 | | Structure of mouse CD1d in complex with C8Ph, different space group | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
3GMM
 
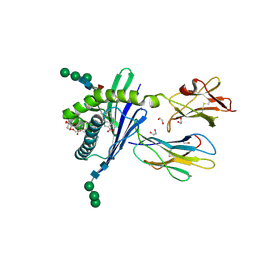 | | Structure of mouse CD1d in complex with C8Ph | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
3GML
 
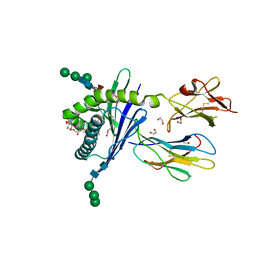 | | Structure of mouse CD1d in complex with C6Ph | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
3GMP
 
 | | Structure of mouse CD1d in complex with PBS-25 | | Descriptor: | (2S,3S,4R)-N-OCTANOYL-1-[(ALPHA-D-GALACTOPYRANOSYL)OXY]-2-AMINO-OCTADECANE-3,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
3GMN
 
 | | Structure of mouse CD1d in complex with C10Ph | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
3HJ0
 
 | |
7JMW
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain in complex with cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
7JMX
 
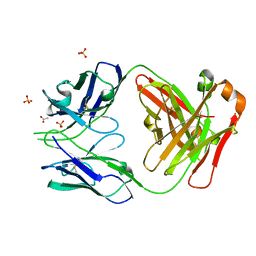 | | Crystal structure of a SARS-CoV-2 cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | ACETATE ION, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
1DBJ
 
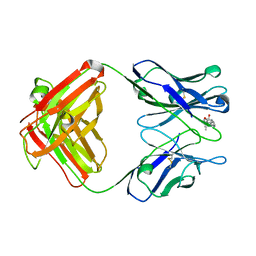 | |
8TFR
 
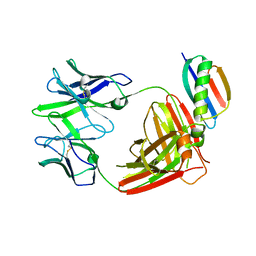 | | Apo Fab from C10-S66K antibody | | Descriptor: | Heavy chain from Fab of C10_S66K antibody, Immunoglobulin G-binding protein G, Light chain from Fab of C10_S66K antibody | | Authors: | Pholcharee, T, Wilson, I.A. | | Deposit date: | 2023-07-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | An Engineered Human-Antibody Fragment with Fentanyl Pan-Specificity That Reverses Carfentanil-Induced Respiratory Depression.
Acs Chem Neurosci, 14, 2023
|
|
8TFP
 
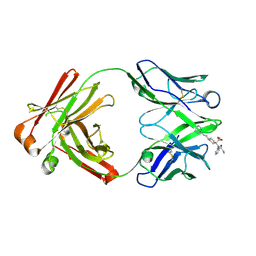 | | Fab from C10-S66K antibody in complex with carfentanil | | Descriptor: | Carfentanil, Heavy chain of Fab from C10-S66K antibody, Light chain of Fab from C10-S66K antibody | | Authors: | Pholcharee, T, Wilson, I.A. | | Deposit date: | 2023-07-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An Engineered Human-Antibody Fragment with Fentanyl Pan-Specificity That Reverses Carfentanil-Induced Respiratory Depression.
Acs Chem Neurosci, 14, 2023
|
|
8TFQ
 
 | | Fab from C10-S66K antibody in complex with fentanyl | | Descriptor: | Heavy chain from Fab of C10_S66K antibody, Light chain from Fab of C10_S66K antibody, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Pholcharee, T, Wilson, I.A. | | Deposit date: | 2023-07-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Human-Antibody Fragment with Fentanyl Pan-Specificity That Reverses Carfentanil-Induced Respiratory Depression.
Acs Chem Neurosci, 14, 2023
|
|
1FRG
 
 | |
1DBK
 
 | |
1DBM
 
 | |
8F0I
 
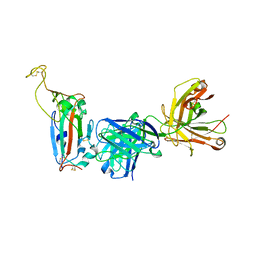 | |
1IAO
 
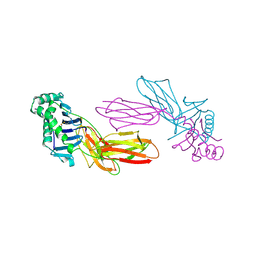 | | CLASS II MHC I-AD IN COMPLEX WITH OVALBUMIN PEPTIDE 323-339 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II I-AD | | Authors: | Scott, C.A, Peterson, P.A, Teyton, L, Wilson, I.A. | | Deposit date: | 1998-03-13 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues.
Immunity, 8, 1998
|
|
2CKB
 
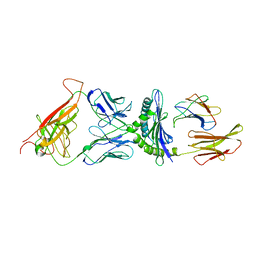 | | STRUCTURE OF THE 2C/KB/DEV8 COMPLEX | | Descriptor: | ALPHA, BETA T CELL RECEPTOR, BETA-2 MICROGLOBULIN, ... | | Authors: | Garcia, K.C, Degano, M, Wilson, I.A. | | Deposit date: | 1998-01-14 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen.
Science, 279, 1998
|
|
2GAR
 
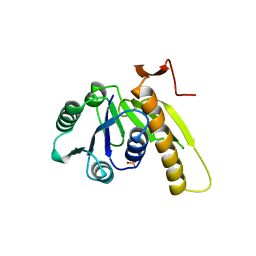 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
2IAD
 
 | |
1C2T
 
 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLASE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-26 | | Release date: | 2000-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
1C3E
 
 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLATE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 2-{4-[2-(2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL)-1-CARBOXY-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-27 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
15C8
 
 | | CATALYTIC ANTIBODY 5C8, FREE FAB | | Descriptor: | IGG 5C8 FAB (HEAVY CHAIN), IGG 5C8 FAB (LIGHT CHAIN) | | Authors: | Gruber, K, Wilson, I.A. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-Induced Conformational Changes in a Catalytic Antibody: Comparison of the Bound and Unbound Structure of Fab 5C8
To be Published
|
|
1GGC
 
 | |
