9BG1
 
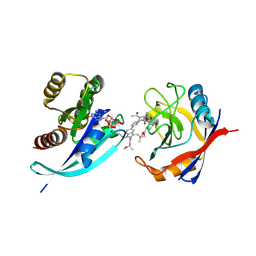 | | Tri-complex of Compound-3, KRAS G12V, and CypA | | Descriptor: | (2R)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1S)-1-methoxyethyl]pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-methyl-2-(N-methylacetamido)butanamide (non-preferred name), GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Tri-complex of Compound-3, KRAS G12V, and CypA
To be published
|
|
9BFY
 
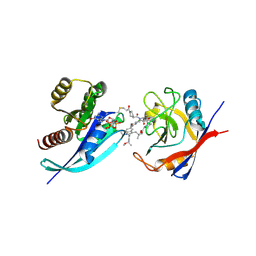 | | Tri-complex of Compound-12, KRAS G12C, and CypA | | Descriptor: | (3R)-N-[(2S)-1-{[(1M,8R,10R,14S,21M)-22-ethyl-4-hydroxy-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]amino}-3-methyl-1-oxobutan-2-yl]-N-methyl-1-propanoylpyrrolidine-3-carboxamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Tri-complex of Compound-12, KRAS G12C, and CypA
To be published
|
|
7BT5
 
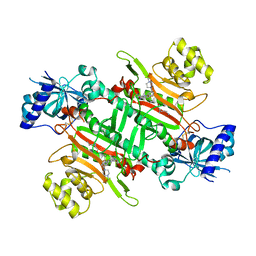 | | Crystal structure of plasmodium LysRS complexing with an antitumor compound | | Descriptor: | LYSINE, Lysine--tRNA ligase, N4-[2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]-N2-(2-propan-2-ylsulfonylphenyl)-1,3,5-triazine-2,4-diamine | | Authors: | Zhou, J, Wang, J, Fang, P. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Inhibition of Plasmodium falciparum Lysyl-tRNA synthetase via an anaplastic lymphoma kinase inhibitor.
Nucleic Acids Res., 48, 2020
|
|
8XLQ
 
 | |
8XLO
 
 | |
7V63
 
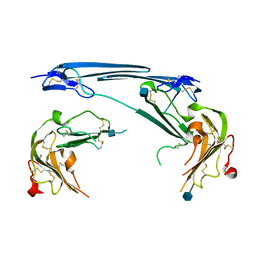 | | Structure of dimeric uPAR at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2021-08-19 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Crystal structure and cellular functions of uPAR dimer
Nat Commun, 13, 2022
|
|
1OQD
 
 | | Crystal structure of sTALL-1 and BCMA | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
1OQE
 
 | | Crystal structure of sTALL-1 with BAFF-R | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
4XFO
 
 | |
4XFN
 
 | |
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2010-08-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
3OQU
 
 | | Crystal structure of native abscisic acid receptor PYL9 with ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL9 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2010-09-04 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
4FHZ
 
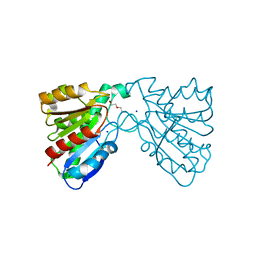 | | Crystal structure of a carboxyl esterase at 2.0 angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phospholipase/Carboxylesterase, SODIUM ION | | Authors: | Wu, L, Ma, J, Zhou, J, Yu, H. | | Deposit date: | 2012-06-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Enhanced enantioselectivity of a carboxyl esterase from Rhodobacter sphaeroides by directed evolution.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
4RIL
 
 | | Structure of the amyloid forming segment, GAVVTGVTAVA, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 68-78, determined by electron diffraction | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.43 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
4JDA
 
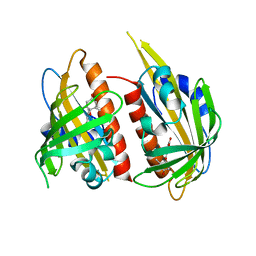 | | Complex structure of abscisic acid receptor PYL3 with (-)-ABA | | Descriptor: | (2Z,4E)-5-[(1R)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Wang, G, Chen, Z. | | Deposit date: | 2013-02-24 | | Release date: | 2013-07-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
4JDL
 
 | |
6C3F
 
 | |
6C88
 
 | |
5H7V
 
 | |
6C3G
 
 | |
4FTW
 
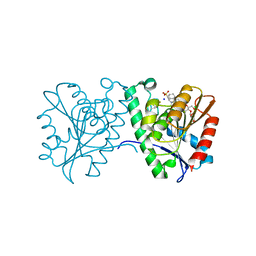 | | Crystal structure of a carboxyl esterase N110C/L145H at 2.3 angstrom resolution | | Descriptor: | 3-CYCLOHEXYLPROPYL 4-O-ALPHA-D-GLUCOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, CHLORIDE ION, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Wu, L, Ma, J, Zhou, J, Yu, H. | | Deposit date: | 2012-06-28 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhanced enantioselectivity of a carboxyl esterase from Rhodobacter sphaeroides by directed evolution.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
4NX2
 
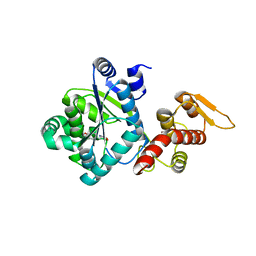 | | Crystal structure of DCYRS complexed with DCY | | Descriptor: | 3,5-dichloro-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Wang, J, Gong, W, Li, J, Gao, F, Li, H. | | Deposit date: | 2013-12-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXE
 
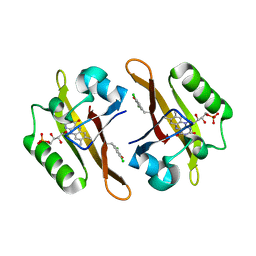 | | Crystal structure of iLOV-I486(2LT) at pH 6.5 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXF
 
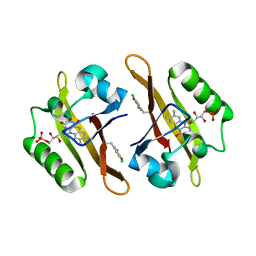 | | Crystal structure of iLOV-I486(2LT) at pH 8.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXG
 
 | | Crystal structure of iLOV-I486z(2LT) at pH 9.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
