8H8F
 
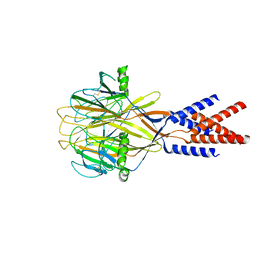 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state) | | Descriptor: | Proton-activated chloride channel | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular insights into the inhibition of proton-activated chloride channel by transfer RNA.
Cell Res., 34, 2024
|
|
8H8E
 
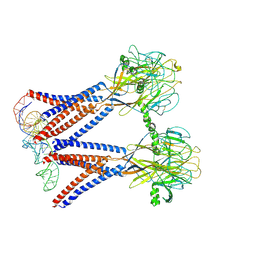 | | Structure of the dimeric Xenopus tropical acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (closed state) | | Descriptor: | Proton-activated chloride channel, tRNA (75-MER)of Spodoptera frugiperda | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Molecular insights into the inhibition of proton-activated chloride channel by transfer RNA.
Cell Res., 34, 2024
|
|
8H8D
 
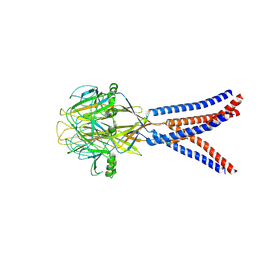 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (intermediate state) | | Descriptor: | Proton-activated chloride channel | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Molecular insights into the inhibition of proton-activated chloride channel by transfer RNA.
Cell Res., 34, 2024
|
|
4FVD
 
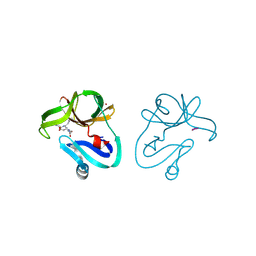 | | Crystal structure of EV71 2A proteinase C110A mutant in complex with substrate | | Descriptor: | 10-mer peptide from 2A proteinase, 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
4FVB
 
 | | Crystal structure of EV71 2A proteinase C110A mutant | | Descriptor: | 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
8H93
 
 | | Structure of dimeric mouse SCMC core complex | | Descriptor: | NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Han, Z, Li, J, Deng, D. | | Deposit date: | 2022-10-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of the subcortical maternal complex and its implications in reproductive disorders.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H94
 
 | | Structure of mouse SCMC bound with KH domain of FILIA | | Descriptor: | NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Han, Z, Li, J, Deng, D. | | Deposit date: | 2022-10-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the subcortical maternal complex and its implications in reproductive disorders.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H96
 
 | | Structure of mouse SCMC core complex | | Descriptor: | NACHT, LRR and PYD domains-containing protein 5, Oocyte-expressed protein homolog, ... | | Authors: | Chi, P, Ou, G, Li, J, Han, Z, Deng, D. | | Deposit date: | 2022-10-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis of the subcortical maternal complex and its implications in reproductive disorders.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HUW
 
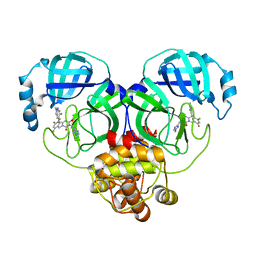 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUX
 
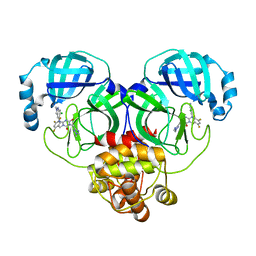 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
4GF6
 
 | | crystal structure of GFP with cuprum bound at the Incorporated metal Chelating Amino Acid PYZ151 | | Descriptor: | CALCIUM ION, COPPER (II) ION, green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4U7P
 
 | | Crystal structure of DNMT3A-DNMT3L complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, L, Guo, X, Li, J, Xiao, J, Yin, X, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.821 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
5DQU
 
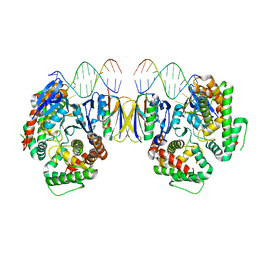 | | Crystal Structure of Cas-DNA-10 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(*GP*AP*GP*TP*CP*GP*AP*TP*GP*CP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
5VGJ
 
 | | Crystal Structure of the Human Fab VRC38.01, an HIV-1 V1V2-Directed Neutralizing Antibody Isolated from Donor N90, bound to a scaffolded WITO V1V2 domain | | Descriptor: | 1FD6-V1V2-WITO, 2-acetamido-2-deoxy-beta-D-glucopyranose, VRC38.01 Fab Heavy Chain, ... | | Authors: | Gorman, J, Li, J, Kwong, P.D. | | Deposit date: | 2017-04-11 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop.
Immunity, 46, 2017
|
|
4U7T
 
 | | Crystal structure of DNMT3A-DNMT3L in complex with histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Guo, X, Wang, L, Yin, X, Li, J, Xiao, J, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
8HUR
 
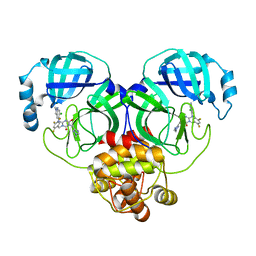 | | Crystal structure of SARS-Cov-2 main protease in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUS
 
 | | Crystal structure of SARS main protease in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUT
 
 | | Crystal structure of MERS main protease in complex with S217622 | | Descriptor: | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ORF1a | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUV
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUU
 
 | | Crystal structure of HCoV-NL63 main protease with S217622 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
5DQT
 
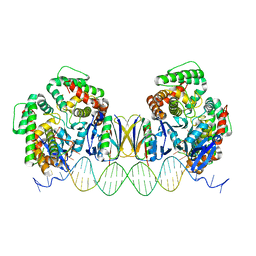 | | Crystal Structure of Cas-DNA-22 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (33-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
3UN0
 
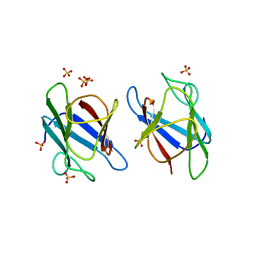 | | Crystal Structure of MDC1 FHA Domain | | Descriptor: | Mediator of DNA damage checkpoint protein 1, SULFATE ION | | Authors: | Clapperton, J.A, Lloyd, J, Haire, L.F, Li, J, Smerdon, S.J. | | Deposit date: | 2011-11-15 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator.
Nucleic Acids Res., 40, 2012
|
|
3UOT
 
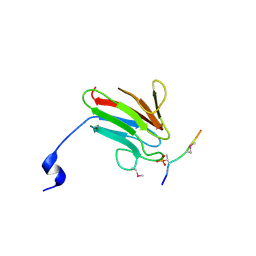 | | Crystal Structure of MDC1 FHA Domain in Complex with a Phosphorylated Peptide from the MDC1 N-terminus | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Clapperton, J.A, Lloyd, J, Haire, L.F, Li, J, Smerdon, S.J. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator.
Nucleic Acids Res., 40, 2012
|
|
6JZO
 
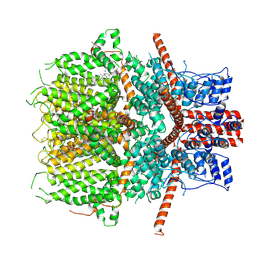 | | Structure of the mouse TRPC4 ion channel | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Duan, J, Li, Z, Li, J, Zhang, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-10-21 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the mouse TRPC4 ion channel
To Be Published
|
|
7C2A
 
 | | Crystal structure of AlinE4, a SGNH-hydrolase family esterase | | Descriptor: | ACETATE ION, CADMIUM ION, SGNH-hydrolase family esterase | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | C-terminal swapped dimers revealed a new catalytic mechanism of SGNH-hydrolase family esterases
To Be Published
|
|
