7XEM
 
 | | Cholesterol bound state of mTRPV2 | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEU
 
 | | Structure of mTRPV2_E2 | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEO
 
 | | MbetaCD treated state of mTRPV2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEV
 
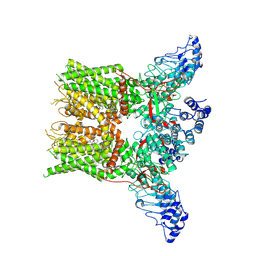 | | Structure of mTRPV2_2-APB | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEW
 
 | | Structure of mTRPV2_Q525F | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XER
 
 | | Structure of mTRPV2_Q525T | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
8YAJ
 
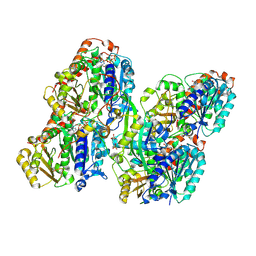 | | ATAT-2 bound MEC-12/MEC-7 microtubule without acetyl-CoA | | Descriptor: | Alpha-tubulin N-acetyltransferase 2, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Lam, W.H, Yu, D, Zhai, Y, Ti, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-11-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Tubulin acetyltransferases access and modify the microtubule luminal K40 residue through anchors in taxane-binding pockets.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8YAR
 
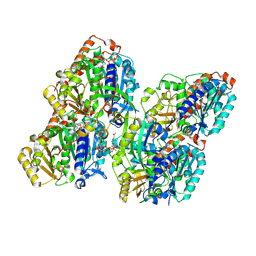 | | ATAT-2 bound K40R MEC-12/MEC-7 microtubule | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lam, W.H, Yu, D, Zhai, Y, Ti, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-11-06 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tubulin acetyltransferases access and modify the microtubule luminal K40 residue through anchors in taxane-binding pockets.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7YEP
 
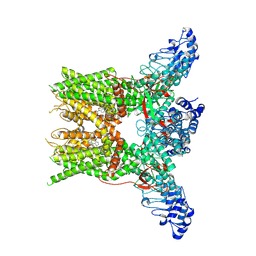 | | 2-APB bound state of mTRPV2 | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-07-06 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7DDX
 
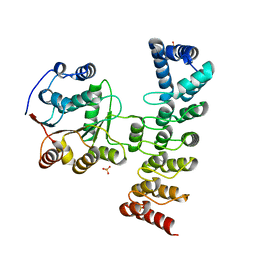 | | Crystal structure of KANK1 S1179F mutant in complex wtih eIF4A1 | | Descriptor: | Eukaryotic initiation factor 4A-I, GLYCEROL, KN motif and ankyrin repeat domains 1, ... | | Authors: | Pan, W, Xu, Y, Wei, Z. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nephrotic-syndrome-associated mutation of KANK2 induces pathologic binding competition with physiological interactor KIF21A.
J.Biol.Chem., 297, 2021
|
|
8IOY
 
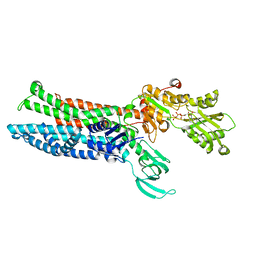 | | Structure of ATP7B C983S/C985S/D1027A mutant with AMP-PNP | | Descriptor: | Copper-transporting ATPase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yang, G, Xu, L, Guo, J, Wu, Z. | | Deposit date: | 2023-03-13 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
7W6S
 
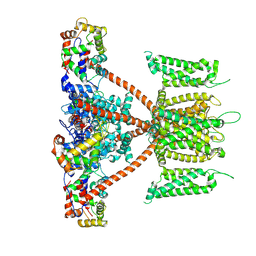 | | CryoEM structure of human KChIP2-Kv4.3 complex | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 2 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7W6N
 
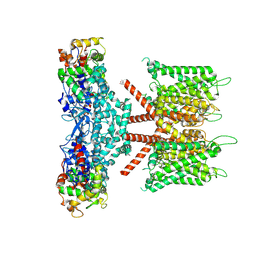 | | CryoEM structure of human KChIP1-Kv4.3 complex | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 1 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7W6T
 
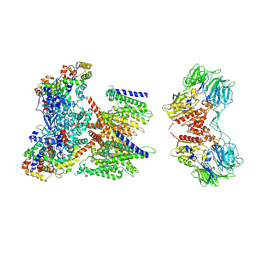 | | CryoEM structure of human KChIP1-Kv4.3-DPP6 complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 1 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7W3Y
 
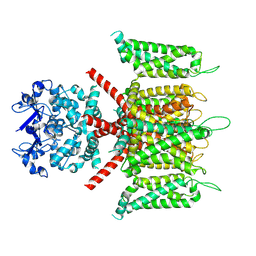 | | CryoEM structure of human Kv4.3 | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7WKS
 
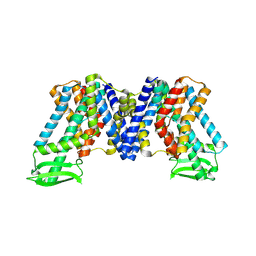 | | Apo state of AtPIN3 | | Descriptor: | Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7WKW
 
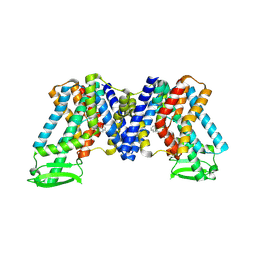 | | NPA bound state of AtPIN3 | | Descriptor: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7V4X
 
 | | Structure of cyclohexanone monooxygenase mutant from Acinetobacter calcoaceticus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H. | | Deposit date: | 2021-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7V50
 
 | | Structure of cyclohexanone monooxygenase mutant from Acinetobacter calcoaceticus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H. | | Deposit date: | 2021-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7V51
 
 | | BVMO_negative mutant D432V | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H.-L. | | Deposit date: | 2021-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7C6S
 
 | |
7CXR
 
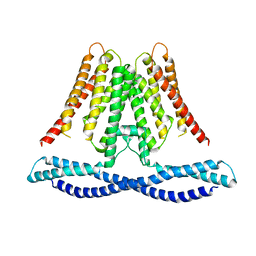 | | Cryo-EM structure of human TMEM120A/TACAN | | Descriptor: | MCherry fluorescent protein,Ion channel TACAN | | Authors: | Yan, Z, Wu, J, Ke, M. | | Deposit date: | 2020-09-02 | | Release date: | 2021-09-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human TMEM120A and TMEM120B.
Cell Discov, 7, 2021
|
|
7BRP
 
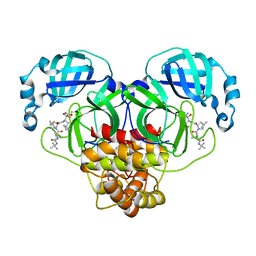 | |
7BRO
 
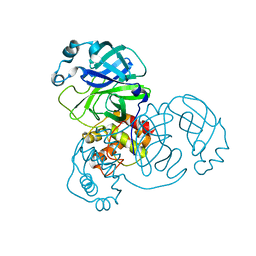 | | Crystal structure of the 2019-nCoV main protease | | Descriptor: | 3C-like proteinase | | Authors: | Fu, L.F. | | Deposit date: | 2020-03-29 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
7D1M
 
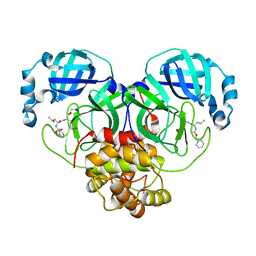 | | CRYSTAL STRUCTURE OF THE SARS-CoV-2 MAIN PROTEASE COMPLEXED WITH GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fu, L.F, Gilski, M, Shabalin, I, Gao, G.F, Qi, J.X. | | Deposit date: | 2020-09-14 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
