6AGH
 
 | | Crystal structure of EFHA1 in Apo-State | | Descriptor: | Calcium uptake protein 2, mitochondrial | | Authors: | Yangfei, X, Xue, Y, Yuequan, S. | | Deposit date: | 2018-08-11 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.742 Å) | | Cite: | Dimerization of MICU Proteins Controls Ca2+Influx through the Mitochondrial Ca2+Uniporter.
Cell Rep, 26, 2019
|
|
6AGJ
 
 | | Crystal Structure of EFHA2 in Apo State | | Descriptor: | Calcium uptake protein 3, mitochondrial | | Authors: | Yangfei, X, Xue, Y, Yuequan, S. | | Deposit date: | 2018-08-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Dimerization of MICU Proteins Controls Ca2+Influx through the Mitochondrial Ca2+Uniporter.
Cell Rep, 26, 2019
|
|
6AGI
 
 | | Crystal Structure of EFHA2 in Ca-binding State | | Descriptor: | CALCIUM ION, Calcium uptake protein 3, mitochondrial, ... | | Authors: | Yangfei, X, Xue, Y, Yuequan, S. | | Deposit date: | 2018-08-11 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Dimerization of MICU Proteins Controls Ca2+Influx through the Mitochondrial Ca2+Uniporter.
Cell Rep, 26, 2019
|
|
5GMQ
 
 | | Structure of MERS-CoV RBD in complex with a fully human antibody MCA1 | | Descriptor: | 1,2-ETHANEDIOL, MCA1 heavy chain, MCA1 light chain, ... | | Authors: | Chen, C, Wang, J.M, Zou, T.T, Gao, X.P, Cui, S, Jin, Q. | | Deposit date: | 2016-07-15 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Human Neutralizing Monoclonal Antibody Inhibition of Middle East Respiratory Syndrome Coronavirus Replication in the Common Marmoset.
J. Infect. Dis., 215, 2017
|
|
5IL1
 
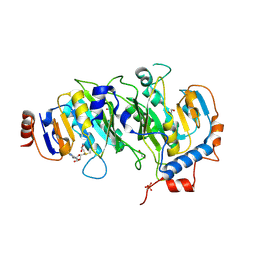 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
2LSJ
 
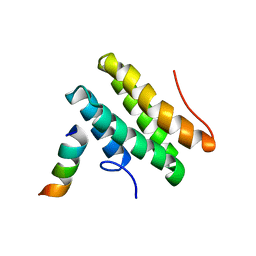 | |
2LSG
 
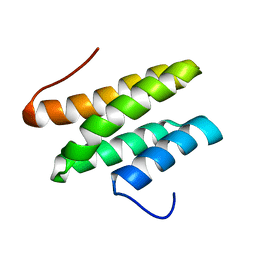 | |
6MEV
 
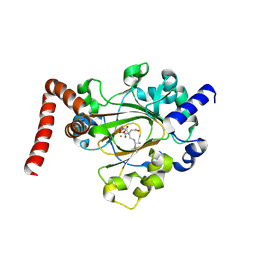 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
2JFV
 
 | |
2JFY
 
 | |
2JFP
 
 | |
2JFZ
 
 | |
2JFW
 
 | |
2JFU
 
 | |
2JFQ
 
 | |
2JFN
 
 | |
7VG3
 
 | | Cryo-EM structure of Arabidopsis DCL3 in complex with a 30-bp RNA | | Descriptor: | CALCIUM ION, Dicer-like 3, TAS1a RNA forward strand (5'-phosphorylated), ... | | Authors: | Wang, Q, Du, J. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation.
Science, 374, 2021
|
|
7VG2
 
 | | Cryo-EM structure of Arabidopsis DCL3 in complex with a 40-bp RNA | | Descriptor: | CALCIUM ION, Dicer-like 3, TAS1a forward strand (5'-phosphorylation), ... | | Authors: | Wang, Q, Du, J. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation.
Science, 374, 2021
|
|
5DDQ
 
 | | L-glutamine riboswitch bound with L-glutamine soaked with Mn2+ | | Descriptor: | GLUTAMINE, L-glutamine riboswitch RNA (61-MER), MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDR
 
 | | L-glutamine riboswitch bound with L-glutamine soaked with Cs+ | | Descriptor: | CESIUM ION, GLUTAMINE, L-glutamine riboswitch RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDP
 
 | | L-glutamine riboswitch bound with L-glutamine | | Descriptor: | GLUTAMINE, MAGNESIUM ION, RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDO
 
 | |
4XOL
 
 | | Observing the overall rocking motion of a protein in a crystal - Cubic Ubiquitin crystals. | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Coquelle, N, Peixiang, M, Schanda, P, Colletier, J.P. | | Deposit date: | 2015-01-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Observing the overall rocking motion of a protein in a crystal.
Nat Commun, 6, 2015
|
|
4XOF
 
 | |
4XOK
 
 | | Observing the overall rocking motion of a protein in a crystal. | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Coquelle, N, Ma, P, Schanda, P, Colletier, J.P. | | Deposit date: | 2015-01-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Observing the overall rocking motion of a protein in a crystal.
Nat Commun, 6, 2015
|
|
