3N52
 
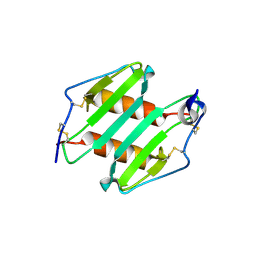 | | crystal Structure analysis of MIP2 | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Rajasekaran, D. | | Deposit date: | 2010-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Model of GAG/MIP-2/CXCR2 Interfaces and Its Functional Effects.
Biochemistry, 51, 2012
|
|
8RFA
 
 | | Arginase 2 in complex with an inhibitor | | Descriptor: | Arginase-2, mitochondrial, MANGANESE (II) ION, ... | | Authors: | Petersen, J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of (2 R ,4 R )-4-(( S )-2-Amino-3-methylbutanamido)-2-(4-boronobutyl)pyrrolidine-2-carboxylic Acid (AZD0011), an Actively Transported Prodrug of a Potent Arginase Inhibitor to Treat Cancer.
J.Med.Chem., 67, 2024
|
|
6J1L
 
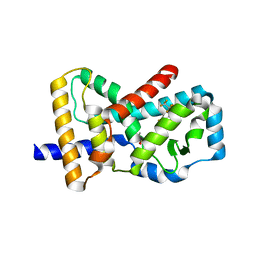 | | Crystal Structure Analysis of the ROR gamma(C455E) | | Descriptor: | 2-[4-(ethylsulfonyl)phenyl]-N-[2'-fluoro-4'-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)[1,1'-biphenyl]-4-yl]acetamide, Nuclear receptor ROR-gamma | | Authors: | zhang, Y, Li, C.C, wu, X.S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Characterization of XY101, a Potent, Selective, and Orally Bioavailable ROR gamma Inverse Agonist for Treatment of Castration-Resistant Prostate Cancer.
J.Med.Chem., 62, 2019
|
|
8YJY
 
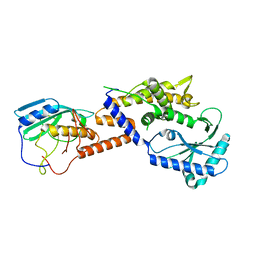 | |
9JJD
 
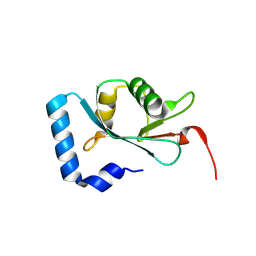 | | Structural analysis of autophagy-related protein 8a in Drosophila melanogaster | | Descriptor: | Autophagy-related 8a, isoform A | | Authors: | Zhang, S.Q, Luo, X, Wu, D.X, Liu, J, Li, X.Y. | | Deposit date: | 2024-09-13 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal Structure of Autophagy-Associated Protein 8 at 1.36 angstrom Resolution and Its Inhibitory Interactions with Indole Analogs.
J.Agric.Food Chem., 73, 2025
|
|
6CN3
 
 | |
6RVV
 
 | | Structure of left-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges. | | Descriptor: | GOLD ION, Transcription attenuation protein MtrB | | Authors: | Malay, A.D, Miyazaki, N, Biela, A.P, Iwasaki, K, Heddle, J.G. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ultra-stable gold-coordinated protein cage displaying reversible assembly.
Nature, 569, 2019
|
|
6CMW
 
 | | Crystal structure of zebrafish Phosphatidylinositol-4-phosphate 5- kinase alpha isoform with bound ATP/Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Phosphatidylinositol-4-phosphate 5-kinase, ... | | Authors: | Zeng, X, Sui, D, Hu, J. | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insights into lethal contractural syndrome type 3 (LCCS3) caused by a missense mutation of PIP5K gamma.
Biochem. J., 475, 2018
|
|
8K0B
 
 | | Cryo-EM structure of TMEM63C | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Qin, Y, Yu, D, Dong, J, Dang, S. | | Deposit date: | 2023-07-08 | | Release date: | 2023-12-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure of TMEM63C suggests it functions as a monomer.
Nat Commun, 14, 2023
|
|
6RVW
 
 | | Structure of right-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges. | | Descriptor: | GOLD ION, Transcription attenuation protein MtrB | | Authors: | Malay, A.D, Miyazaki, N, Biela, A.P, Iwasaki, K, Heddle, J.G. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ultra-stable gold-coordinated protein cage displaying reversible assembly.
Nature, 569, 2019
|
|
7N9L
 
 | | KirBac3.1 C71S C262S | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Inward rectifier potassium channel Kirbac3.1, POTASSIUM ION, ... | | Authors: | Gulbis, J.M, Black, K.A. | | Deposit date: | 2021-06-18 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ion currents through Kir potassium channels are gated by anionic lipids.
Nat Commun, 13, 2022
|
|
5IWL
 
 | | CD47-diabody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5F9 diabody, Leukocyte surface antigen CD47, ... | | Authors: | Di, W, Jude, K.M, Garcia, K.C. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer.
J.Clin.Invest., 126, 2016
|
|
6CN2
 
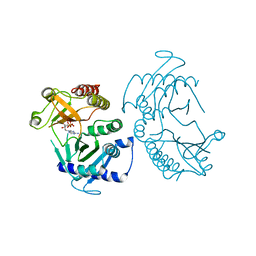 | | Crystal structure of zebrafish Phosphatidylinositol-4-phosphate 5- kinase alpha isoform D236N with bound ATP/Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Phosphatidylinositol-4-phosphate 5-kinase, ... | | Authors: | Zeng, X, Sui, D, Hu, J. | | Deposit date: | 2018-03-07 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural insights into lethal contractural syndrome type 3 (LCCS3) caused by a missense mutation of PIP5K gamma.
Biochem. J., 475, 2018
|
|
7N9K
 
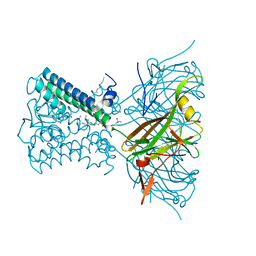 | | KirBac3.1 L124M mutant | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Inward rectifier potassium channel Kirbac3.1, N-OCTANE, ... | | Authors: | Black, T.A, Gulbis, J.M. | | Deposit date: | 2021-06-18 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Ion currents through Kir potassium channels are gated by anionic lipids.
Nat Commun, 13, 2022
|
|
7CCH
 
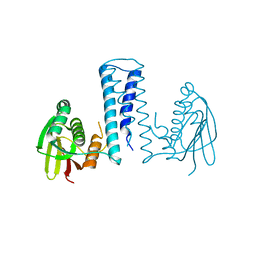 | | Acinetobacter baumannii histidine kinase AdeS | | Descriptor: | AdeS | | Authors: | Wen, Y, Felix, J. | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.848 Å) | | Cite: | Proteolysis and multimerization regulate signaling along the two-component regulatory system AdeRS.
Iscience, 24, 2021
|
|
7CCI
 
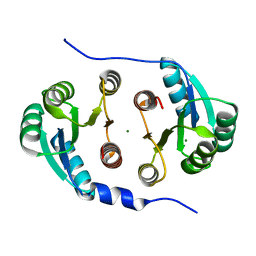 | |
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
9J7V
 
 | | Human G6PC1 in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glucose-6-phosphatase catalytic subunit 1 | | Authors: | Jiang, D.H, Xia, Z.Y. | | Deposit date: | 2024-08-19 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into glucose-6-phosphate recognition and hydrolysis by human G6PC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9J7U
 
 | | H176A mutant of human G6PC1 in complex with G6P | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-O-phosphono-beta-D-glucopyranose, Glucose-6-phosphatase catalytic subunit 1 | | Authors: | Jiang, D.H, Xia, Z.Y. | | Deposit date: | 2024-08-19 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural insights into glucose-6-phosphate recognition and hydrolysis by human G6PC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6OQT
 
 | | E. coli ATP synthase State 1c | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6IJX
 
 | | Crystal Structure of AKR1C1 complexed with meclofenamic acid | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, L, Zhang, H, Chen, Y, Hu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
6PQV
 
 | | E. coli ATP Synthase State 1e | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-07-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6OQS
 
 | | E. coli ATP synthase State 1b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6OQW
 
 | | E. coli ATP synthase State 3a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-24 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6A7B
 
 | | AKR1C3 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C3, DIMETHYLFORMAMIDE, ... | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
