1KLO
 
 | |
3AQE
 
 | | Crystal structure of the extracellular domain of human RAMP2 | | Descriptor: | Receptor activity-modifying protein 2 | | Authors: | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Shindo, T, Yokoyama, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-09 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding
Protein Sci., 21, 2012
|
|
3AQF
 
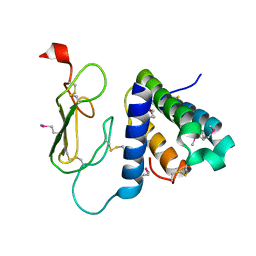 | | Crystal structure of the human CRLR/RAMP2 extracellular complex | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Receptor activity-modifying protein 2 | | Authors: | Kusano, S, Kukimono-Niino, M, Shirouzu, M, Shindo, T, Yokoyama, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding
Protein Sci., 21, 2012
|
|
7D2O
 
 | |
1TLE
 
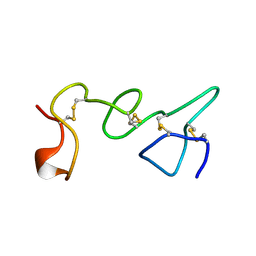 | | LE (LAMININ-TYPE EGF-LIKE) MODULE GIII4 IN SOLUTION AT PH 3.5 AND 290 K, NMR, 14 STRUCTURES | | Descriptor: | LAMININ | | Authors: | Baumgartner, R, Czisch, M, Mayer, U, Schl, E.P, Huber, R, Timpl, R, Holak, T.A. | | Deposit date: | 1996-01-26 | | Release date: | 1997-02-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the nidogen binding LE module of the laminin gamma1 chain in solution.
J.Mol.Biol., 257, 1996
|
|
7WUX
 
 | | Crystal structure of AziU3/U2 complexed with (5S,6S)-O7-sulfo DADH from Streptomyces sahachiroi | | Descriptor: | (2S,5S,6S)-2,6-bis(azanyl)-5-oxidanyl-7-sulfooxy-heptanoic acid, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, AziU2, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
7WUW
 
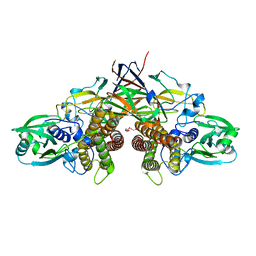 | | Crystal structure of AziU3/U2 from Streptomyces sahachiroi | | Descriptor: | AziU2, AziU3, MAGNESIUM ION, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
2C32
 
 | |
2ELL
 
 | | Solution structure of the Leucine Rich Repeat of human Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Authors: | Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
2AYQ
 
 | |
2ZJ9
 
 | | X-ray crystal structure of AmpC beta-Lactamase (AmpC(D)) from an Escherichia coli with a Tripeptide Deletion (Gly286 Ser287 Asp288) on the H10 Helix | | Descriptor: | AmpC, ISOPROPYL ALCOHOL, SODIUM ION | | Authors: | Yamaguchi, Y, Sato, G, Yamagata, Y, Wachino, J, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2008-02-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of AmpC beta-lactamase (AmpCD) from an Escherichia coli clinical isolate with a tripeptide deletion (Gly286-Ser287-Asp288) in the H10 helix
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1JJ0
 
 | |
1JIT
 
 | |
1JJ3
 
 | |
1JIS
 
 | |
1JIY
 
 | |
1JJ1
 
 | |
