3EUH
 
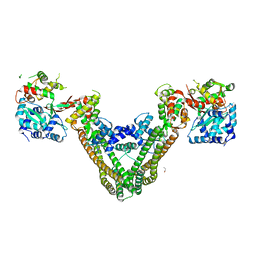 | | Crystal Structure of the MukE-MukF Complex | | Descriptor: | Chromosome partition protein mukF, GLYCINE, MukE | | Authors: | Suh, M.K, Ku, B, Ha, N.C, Woo, J.S, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
2JXB
 
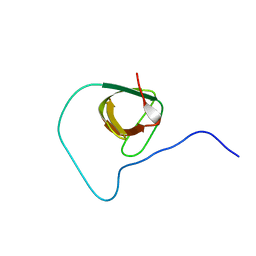 | | Structure of CD3epsilon-Nck2 first SH3 domain complex | | Descriptor: | T-cell surface glycoprotein CD3 epsilon chain, Cytoplasmic protein NCK2 | | Authors: | Takeuchi, K, Yang, H, Ng, E, Park, S, Sun, Z.J, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional evidence that Nck interaction with CD3epsilon regulates T-cell receptor activity.
J.Mol.Biol., 380, 2008
|
|
3FLM
 
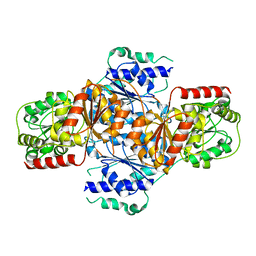 | | Crystal structure of menD from E.coli | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Priyadarshi, A, Hwang, K.Y. | | Deposit date: | 2008-12-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights of the MenD from Escherichia coli reveal ThDP affinity.
Biochem.Biophys.Res.Commun., 380, 2009
|
|
4LW4
 
 | |
4LW2
 
 | |
3QN1
 
 | | Crystal structure of the PYR1 Abscisic Acid receptor in complex with the HAB1 type 2C phosphatase catalytic domain | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Marquez, J.A. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of Abscisic Acid Signaling in Vivo by an Engineered Receptor-Insensitive Protein Phosphatase Type 2C Allele.
Plant Physiol., 156, 2011
|
|
1GV2
 
 | | CRYSTAL STRUCTURE OF C-MYB R2R3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-05 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
3V7A
 
 | |
5HXY
 
 | | Crystal structure of XerA recombinase | | Descriptor: | PHOSPHATE ION, Tyrosine recombinase XerA | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Thermoplasma acidophilum XerA recombinase shows large C-shape clamp conformation and cis-cleavage mode for nucleophilic tyrosine
FEBS Lett., 590, 2016
|
|
4LA7
 
 | | X-ray crystal structure of the PYL2-quinabactin-Hab1 ternary complex | | Descriptor: | ACETATE ION, Abscisic acid receptor PYL2, GLYCEROL, ... | | Authors: | Peterson, F.C, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5I1Z
 
 | | Structure of nvPizza2-H16S58 | | Descriptor: | SULFATE ION, nvPizza2-H16S58 | | Authors: | Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Broken Symmetry: Partial domain swapping in an artificial trimeric protein
To Be Published
|
|
6KZ7
 
 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | Descriptor: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | Deposit date: | 2019-09-23 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
5ZJE
 
 | | LDHA-mla | | Descriptor: | L-lactate dehydrogenase A chain, MALONATE ION | | Authors: | Han, C.W, Jang, S.B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.929 Å) | | Cite: | Machilin A Inhibits Tumor Growth and Macrophage M2 Polarization Through the Reduction of Lactic Acid.
Cancers (Basel), 11, 2019
|
|
5ZJF
 
 | | LDHA-MA | | Descriptor: | 5,5'-[(2R,3S)-2,3-dimethylbutane-1,4-diyl]bis(2H-1,3-benzodioxole), L-lactate dehydrogenase A chain | | Authors: | Han, C.W, Jang, S.B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Machilin A Inhibits Tumor Growth and Macrophage M2 Polarization Through the Reduction of Lactic Acid.
Cancers (Basel), 11, 2019
|
|
5ZJD
 
 | | Lactate dehydrogenase with NADH and MLA | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase A chain, MALONATE ION | | Authors: | Han, C.W, Jang, S.B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Machilin A Inhibits Tumor Growth and Macrophage M2 Polarization Through the Reduction of Lactic Acid.
Cancers (Basel), 11, 2019
|
|
7CRJ
 
 | | Dark State Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRL
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 50 ps after light activation | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRY
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (6.49 mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRK
 
 | | 2ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRX
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (2.63mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRS
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.90mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2HJW
 
 | | Crystal Structure of the BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2006-07-02 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the biotin carboxylase domain of human acetyl-CoA carboxylase 2.
Proteins, 70, 2008
|
|
7CRI
 
 | | 1 ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DVD
 
 | | The crystal structure of p53 DNA binding domain and PUMA complex | | Descriptor: | Bcl-2-binding component 3, isoforms 1/2, Cellular tumor antigen p53, ... | | Authors: | Han, C.W, Lee, H.N, Jeong, M.S, Jang, S.B. | | Deposit date: | 2021-01-13 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis of the p53 DNA binding domain and PUMA complex.
Biochem.Biophys.Res.Commun., 548, 2021
|
|
