1XGF
 
 | |
5X7K
 
 | |
3E3B
 
 | |
1TEZ
 
 | | COMPLEX BETWEEN DNA AND THE DNA PHOTOLYASE FROM ANACYSTIS NIDULANS | | Descriptor: | 5'-D(*AP*TP*CP*GP*GP*CP*T*(TCP)P*CP*GP*C)-3', 5'-D(*TP*CP*GP*C)-3', 5'-D(P*CP*GP*AP*AP*GP*CP*CP*GP*A)-3', ... | | Authors: | Essen, L.-O, Carell, T, Mees, A, Klar, T. | | Deposit date: | 2004-05-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair
Science, 306, 2004
|
|
2DC6
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA073 complex | | Descriptor: | BENZYL N-({(2S,3S)-3-[(PROPYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-28 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DC7
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA042 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-THREONYL-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCC
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA077 complex | | Descriptor: | BENZYL N-({(2S,3S)-3-[(BENZYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, CATHEPSIN B, GLYCEROL, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCD
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA078 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-({(2S,3S)-3-[(BENZYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCB
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA076 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCYL-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCA
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA075 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCYL-L-ALANINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DC9
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA074Me complex | | Descriptor: | CATHEPSIN B, GLYCEROL, METHYL N-({(2S,3S)-3-[(PROPYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DC8
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA059 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2ZZO
 
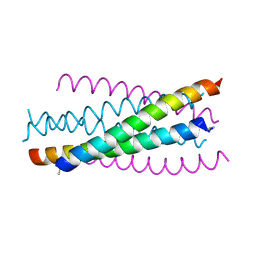 | | Crystal structure of the complex between GP41 fragment N36 and fusion inhibitor C34/S138A | | Descriptor: | Transmembrane protein | | Authors: | Watabe, T, Nakano, H, Nakatsu, T, Kato, H, Fujii, N. | | Deposit date: | 2009-02-20 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic study of an HIV-1 fusion inhibitor with the gp41 S138A substitution
J.Mol.Biol., 392, 2009
|
|
2DKV
 
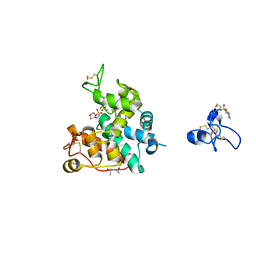 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, chitinase | | Authors: | Kezuka, Y, Nishizawa, Y, Watanabe, T, Nonaka, T. | | Deposit date: | 2006-04-14 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
3ABQ
 
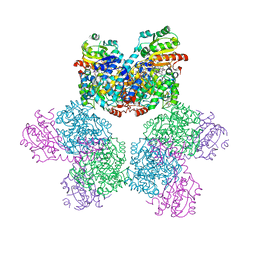 | |
3ABR
 
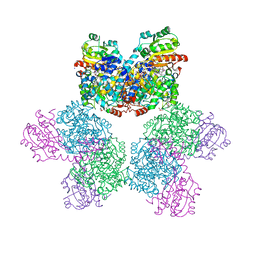 | |
3ABO
 
 | |
3ABS
 
 | |
1KUH
 
 | | ZINC PROTEASE FROM STREPTOMYCES CAESPITOSUS | | Descriptor: | CALCIUM ION, ZINC ION, ZINC PROTEASE | | Authors: | Kurisu, G, Kinoshita, T, Sugimoto, A, Nagara, A, Kai, Y, Kasai, N, Harada, S. | | Deposit date: | 1996-02-22 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the zinc endoprotease from Streptomyces caespitosus.
J.Biochem.(Tokyo), 121, 1997
|
|
1KMF
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALLO-ILE, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Xu, B, Hua, Q.X, Nakagawa, S.H, Jia, W, Chu, Y.C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-09 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Chiral mutagenesis of insulin's hidden receptor-binding surface: structure of an allo-isoleucine(A2) analogue.
J.Mol.Biol., 316, 2002
|
|
3A12
 
 | | Crystal structure of Type III Rubisco complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
2ZEQ
 
 | | Crystal structure of ubiquitin-like domain of murine Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin | | Authors: | Tomoo, K. | | Deposit date: | 2007-12-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and molecular dynamics simulation of ubiquitin-like domain of murine parkin
Biochim.Biophys.Acta, 1784, 2008
|
|
3AXW
 
 | |
2DQ7
 
 | |
3AT4
 
 | | Crystal structure of CK2alpha with pyradine derivertive | | Descriptor: | Casein kinase II subunit alpha, [1-(6-{6-[(1-methylethyl)amino]-1H-indazol-1-yl}pyrazin-2-yl)-1H-pyrrol-3-yl]acetic acid | | Authors: | Kinoshita, T. | | Deposit date: | 2010-12-23 | | Release date: | 2011-11-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A detailed thermodynamic profile of cyclopentyl and isopropyl derivatives binding to CK2 kinase
Mol.Cell.Biochem., 356, 2011
|
|
