8ZFE
 
 | |
8ZFA
 
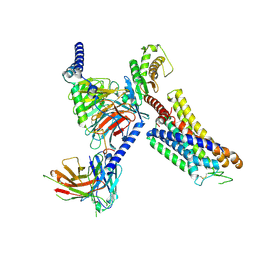 | | Cryo-EM structure of the xtGPR4-Gs complex in pH7.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZF9
 
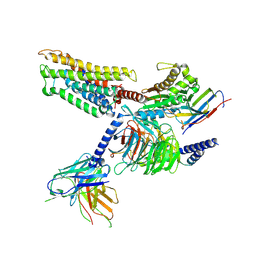 | | Cryo-EM structure of the mmGPR4-Gs complex in pH7.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wen, X, Rong, N.K, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZD1
 
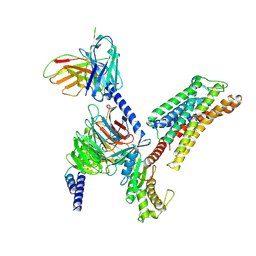 | | Cryo-EM structure of the xGPR4-Gs complex in pH6.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | Deposit date: | 2024-04-30 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZF6
 
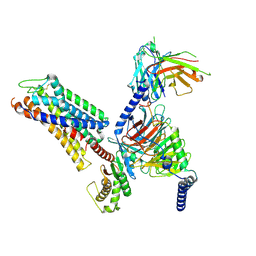 | | Cryo-EM structure of the xGPR4-Gs complex in pH6.7 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Wen, X, Yang, F, Sun, J.P. | | Deposit date: | 2024-05-07 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
8ZFD
 
 | |
5H19
 
 | | EED in complex with PRC2 allosteric inhibitor EED162 | | Descriptor: | 5-(furan-2-ylmethylamino)-9-(phenylmethyl)-8,10-dihydro-7H-[1,2,4]triazolo[3,4-a][2,7]naphthyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
7EE5
 
 | | Crystal structure of Neu5Gc bound PltC | | Descriptor: | N-glycolyl-alpha-neuraminic acid, N-glycolyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, Subtilase cytotoxin subunit B-like protein, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE4
 
 | | Crystal structure of Neu5Ac bound PltC | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE6
 
 | | Crystal structure of PltC toxin | | Descriptor: | ACETONE, CITRATE ANION, Cytolethal distending toxin subunit B family protein, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE3
 
 | | Crystal structure of PltC | | Descriptor: | Subtilase cytotoxin subunit B-like protein, TETRAETHYLENE GLYCOL | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
8JIY
 
 | | A carbohydrate binding domain of a putative chondroitinase | | Descriptor: | DUF4955 domain-containing protein | | Authors: | Liu, G.C, Chang, Y.G. | | Deposit date: | 2023-05-29 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and structural characterization of a novel chondroitin sulfate-specific carbohydrate-binding module: The first member of a new family, CBM100.
Int.J.Biol.Macromol., 255, 2024
|
|
8JCN
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 58 | | Descriptor: | 1-[3-(diphenoxyphosphorylamino)phenyl]ethanone, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCK
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 32 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCM
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 55 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, HYDROSULFURIC ACID, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCO
 
 | |
8JCL
 
 | |
8JCJ
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 18 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
7Y3E
 
 | |
5WBH
 
 | |
5WBU
 
 | |
5WBY
 
 | |
8HYA
 
 | |
8XIF
 
 | | The crystal structure of the AEP domain of VACV D5 | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, Uncoating factor OPG117 | | Authors: | Gan, J, Zhang, W. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8XJ8
 
 | | The Cryo-EM structure of MPXV E5 C-terminal in complex with DNA | | Descriptor: | DNA (70-MER), MAGNESIUM ION, Monkeypox virus E5, ... | | Authors: | Zhang, W, Liu, Y, Gao, H, Gan, J. | | Deposit date: | 2023-12-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
