8EF5
 
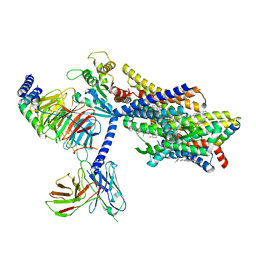 | | Fentanyl-bound mu-opioid receptor-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFO
 
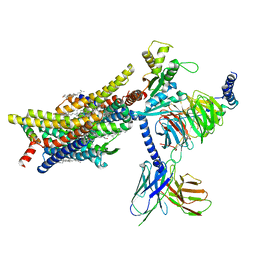 | | PZM21-bound mu-opioid receptor-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EF6
 
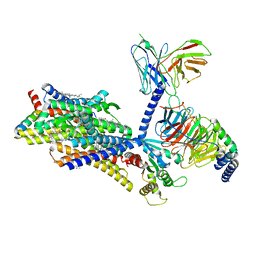 | | Morphine-bound mu-opioid receptor-Gi complex | | Descriptor: | (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFB
 
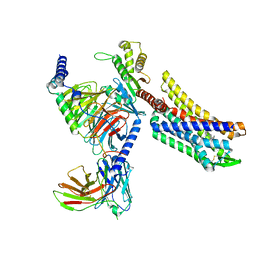 | | Oliceridine-bound mu-opioid receptor-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFL
 
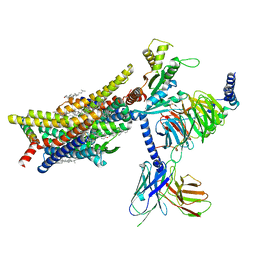 | | SR17018-bound mu-opioid receptor-Gi complex | | Descriptor: | 5,6-dichloro-1-{1-[(4-chlorophenyl)methyl]piperidin-4-yl}-1,3-dihydro-2H-benzimidazol-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFQ
 
 | | DAMGO-bound mu-opioid receptor-Gi complex | | Descriptor: | DAMGO, ETHANOLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
9UDE
 
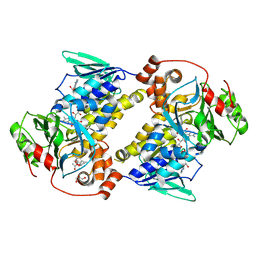 | | Crystal structure of MonCI in complex with diepoxidized farnesyl acetate | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xiao, H.L, Guan, Y.Z, Chen, X. | | Deposit date: | 2025-04-06 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
9UDB
 
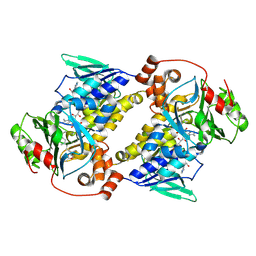 | | Crystal structure of MonCI in complex with farnesyl acetate | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl acetate, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Deng, Y.M, Chen, X. | | Deposit date: | 2025-04-06 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
9UDD
 
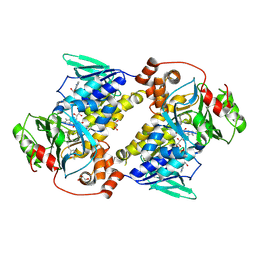 | | Crystal structure of MonCI in complex with monoepoxidized farnesyl acetate | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xiao, H.L, Guan, Y.Z, Chen, X. | | Deposit date: | 2025-04-06 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
8E3D
 
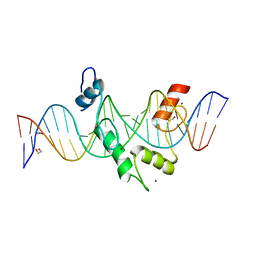 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing CAST sequence (#11) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*TP*TP*GP*GP*GP*GP*AP*GP*GP*GP*GP*TP*CP*TP*TP*TP*AP*AP*CP*C)-3'), DNA (5'-D(P*GP*GP*TP*AP*AP*AP*AP*GP*AP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*AP*AP*T)-3'), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
8E3E
 
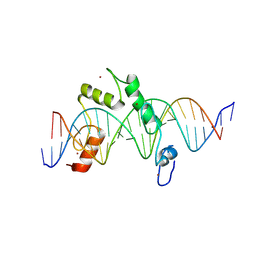 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing CAST sequence (#10) | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*GP*GP*GP*GP*AP*GP*GP*GP*GP*TP*CP*TP*TP*TP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*AP*AP*AP*GP*AP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*AP*A)-3'), ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
8F8Y
 
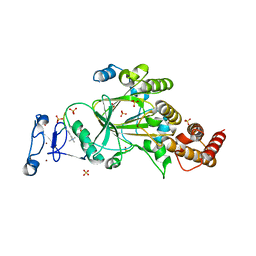 | | PHF2 (PHD+JMJ) in Complex with VRK1 N-Terminal Peptide | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase PHF2, SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2022-11-22 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A complete methyl-lysine binding aromatic cage constructed by two domains of PHF2.
J.Biol.Chem., 299, 2022
|
|
8F8Z
 
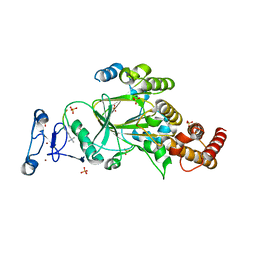 | |
9LXO
 
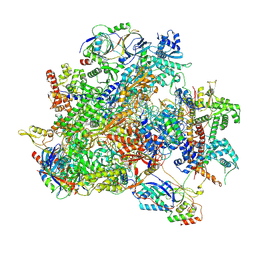 | | Human RNA Polymerase III de novo transcribing complex 6 (TC6)(without 3'dATP) | | Descriptor: | DNA (127-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2025-02-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
9LXN
 
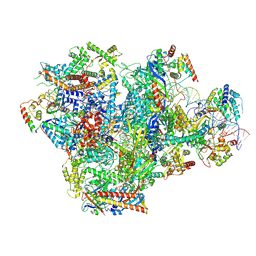 | | Human RNA Polymerase III de novo transcribing complex 5 (TC5)(without 3'dATP) | | Descriptor: | DNA (127-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Wang, Q, Ren, Y, Jin, Q, Chen, X, Xu, Y. | | Deposit date: | 2025-02-18 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into human Pol III transcription initiation in action.
Nature, 2025
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | Descriptor: | CidA_I gamma/2 protein, CidB_I b/2 protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | Descriptor: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | Descriptor: | ULP_PROTEASE domain-containing protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | Descriptor: | bacteria factor 1 | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
4AY9
 
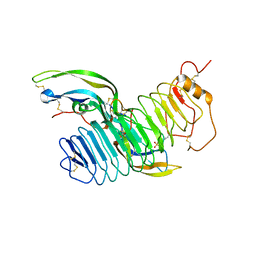 | | Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLLICLE-STIMULATING HORMONE RECEPTOR, FOLLITROPIN SUBUNIT BETA, ... | | Authors: | Jiang, X, Liu, H, Chen, X, He, X. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Follicle-Stimulating Hormone in Complex with the Entire Ectodomain of its Receptor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6DQB
 
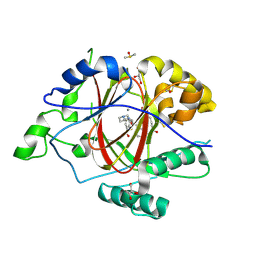 | | LINKED KDM5A JMJ DOMAIN FORMING COVALENT BOND TO INHIBITOR N71 i.e. 2-((3-(4-(dimethylamino)but-2-enamido)phenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 2-{(R)-(3-{[(2E)-4-(dimethylamino)but-2-enoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(R)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(S)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ6
 
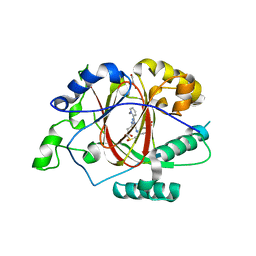 | |
6DQA
 
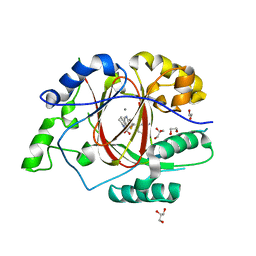 | | Linked KDM5A JMJ Domain Bound to Inhibitor N70 i.e.[2-((3-aminophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-(3-aminophenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ9
 
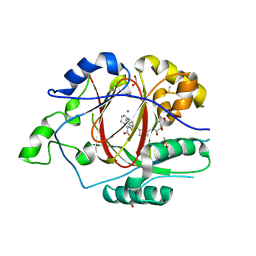 | | Linked KDM5A JMJ Domain Bound to the Covalent Inhibitor N69 i.e. [2-((3-acrylamidophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-[3-(acryloylamino)phenyl][2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ5
 
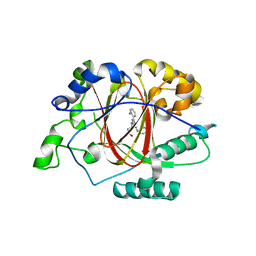 | |
