8YZ7
 
 | |
6KZD
 
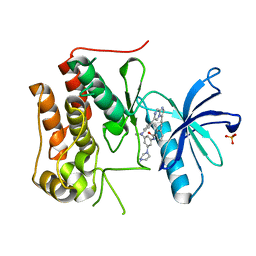 | | Crystal structure of TRKc in complex with 3-((6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl)ethynyl)- N-(3-isopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-2- methylbenzamide | | Descriptor: | 3-[2-[6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl]ethynyl]-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-propan-2-yl-phenyl]benzamide, NT-3 growth factor receptor, PHOSPHATE ION | | Authors: | Wang, Y, Zhang, Z.M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
8HXB
 
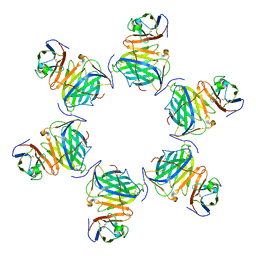 | | Cryo-EM structure of MPXV M2 hexamer in complex with human B7.2 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD86 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8HXC
 
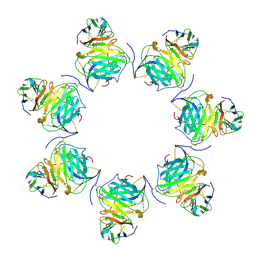 | | Cryo-EM structure of MPXV M2 heptamer in complex with human B7.2 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD86 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8HXA
 
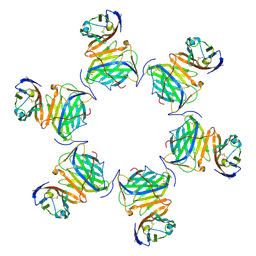 | | Cryo-EM structure of MPXV M2 in complex with human B7.1 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD80 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
6AK4
 
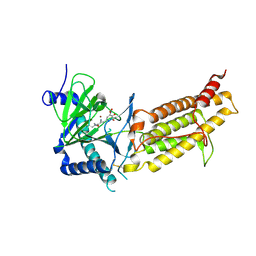 | | Crystal structure of human FTO in complex with small-molecule inhibitors | | Descriptor: | (~{E})-2-cyano-~{N},~{N}-diethyl-3-[3-nitro-4,5-bis(oxidanyl)phenyl]prop-2-enamide, Alpha-ketoglutarate-dependent dioxygenase FTO,Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Wang, Y, Cao, R, Peng, S, Zhang, W, Huang, N. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1.
Sci Transl Med, 11, 2019
|
|
1FB9
 
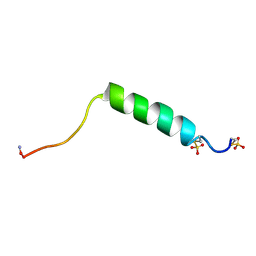 | |
8SPP
 
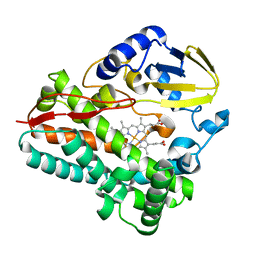 | |
8XAU
 
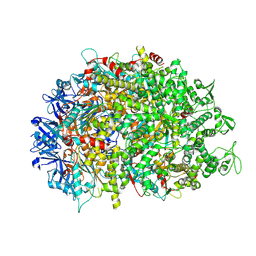 | | Cryo-EM structure of HerA | | Descriptor: | ATP-binding protein | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 2024
|
|
8XAV
 
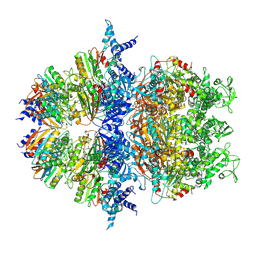 | | Cryo-EM structure of an anti-phage defense complex | | Descriptor: | ATP-binding protein, DUF4297 | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 2024
|
|
6LYH
 
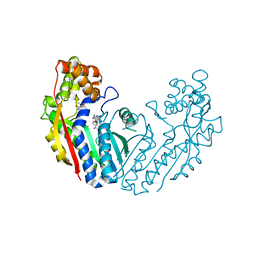 | | Crystal structure of tea N9-methyltransferase CkTcS in complex with SAH and 1,3,7-trimethyluric acid | | Descriptor: | 1,3,7-trimethyl-9H-purine-2,6,8-trione, N-methyltransferase CkTcS, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wang, Y, Zhang, Z.-M. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15000319 Å) | | Cite: | Identification and characterization of N9-methyltransferase involved in converting caffeine into non-stimulatory theacrine in tea.
Nat Commun, 11, 2020
|
|
6LYI
 
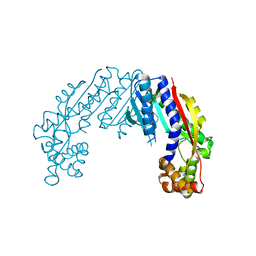 | |
5H1K
 
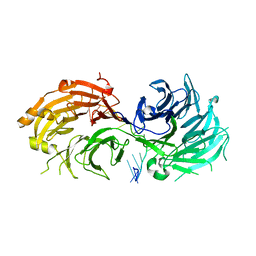 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 13-nt U4 snRNA fragment | | Descriptor: | Gem-associated protein 5, U4 snRNA (5'-R(*GP*CP*AP*AP*UP*UP*UP*UP*UP*GP*AP*CP*A)-3') | | Authors: | Wang, Y, Jin, W, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
139D
 
 | |
7DAD
 
 | | EPD in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y, Wu, C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | High-resolution X-ray structure of three microtubule-stabilizing agents in complex with tubulin provide a rationale for drug design.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7CE2
 
 | | The Crystal structure of TeNT Hc complexed with neutralizing antibody | | Descriptor: | Tetanus toxin, neutralizing antibody heavy chain, neutralizing antibody light chain | | Authors: | Wang, X, Wang, Y, Wu, C, Yu, J, Liao, H. | | Deposit date: | 2020-06-21 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of tetanus toxin neutralization by native human monoclonal antibodies.
Cell Rep, 35, 2021
|
|
7PEM
 
 | | Cryo-EM structure of phophorylated Drs2p-Cdc50p in a PS and ATP-bound E2P state | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Timcenko, M, Wang, Y, Lyons, J.A, Nissen, P, Lindorff-Larsen, K. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Substrate Transport and Specificity in a Phospholipid Flippase
To Be Published
|
|
5K13
 
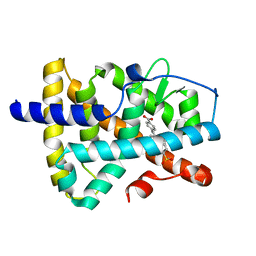 | | Crystal structure of the RAR alpha ligand-binding domain in complex with an antagonist | | Descriptor: | 4-{5-(3-tert-butylphenyl)-1-[4-(methylsulfonyl)phenyl]-1H-pyrazol-3-yl}benzoic acid, Retinoic acid receptor alpha | | Authors: | Wang, Y, Stout, S.L. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of potent and selective retinoic acid receptor gamma (RAR gamma ) antagonists for the treatment of osteoarthritis pain using structure based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8HN9
 
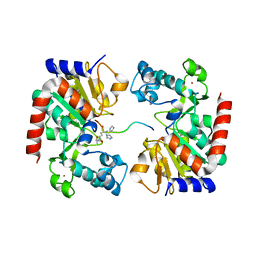 | | Human SIRT3 Recognizing CCNE2K348la peptide | | Descriptor: | CCNE2 peptide, IMIDAZOLE, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Wang, Y, Ding, W. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | SIRT3-dependent delactylation of cyclin E2 prevents hepatocellular carcinoma growth.
Embo Rep., 24, 2023
|
|
5IY4
 
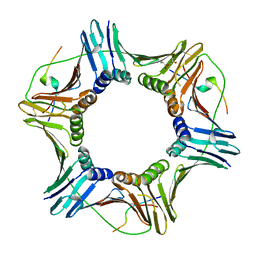 | |
3NYL
 
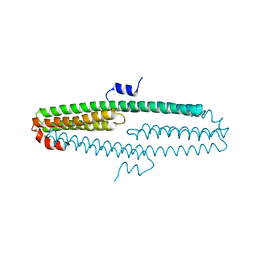 | | The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain | | Descriptor: | Amyloid beta (A4) protein (Peptidase nexin-II, Alzheimer disease), isoform CRA_b | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X, Wang, Y. | | Deposit date: | 2010-07-15 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain.
Mol.Cell, 15, 2004
|
|
5D8Q
 
 | | 2.20A resolution structure of BfrB (L68A) from Pseudomonas aeruginosa | | Descriptor: | ARSENIC, Ferroxidase, MAGNESIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
5D8P
 
 | | 2.35A resolution structure of iron bound BfrB (wild-type, C2221 form) from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
5D8S
 
 | | 2.55A resolution structure of BfrB (E85A) from Pseudomonas aeruginosa | | Descriptor: | Ferroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
5D8R
 
 | | 2.50A resolution structure of BfrB (E81A) from Pseudomonas aeruginosa | | Descriptor: | Ferroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
