8IQB
 
 | |
8IQD
 
 | |
7MQS
 
 | | The insulin receptor ectodomain in complex with three venom hybrid insulin molecules - asymmetric conformation | | Descriptor: | Insulin A chain, Insulin B chain, Isoform Short of Insulin receptor | | Authors: | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | Deposit date: | 2021-05-06 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
7MQR
 
 | | The insulin receptor ectodomain in complex with four venom hybrid insulins - symmetric conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | Authors: | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | Deposit date: | 2021-05-06 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
7MQO
 
 | | The insulin receptor ectodomain in complex with a venom hybrid insulin analog - "head" region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, ... | | Authors: | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | Deposit date: | 2021-05-06 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
8XL3
 
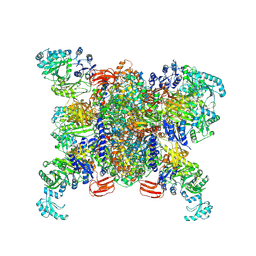 | | Structure of human propionyl-CoA carboxylase at apo-state (PCC-Apo) | | Descriptor: | BIOTIN, Propionyl-CoA carboxylase alpha chain, mitochondrial, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
7WRY
 
 | |
6W44
 
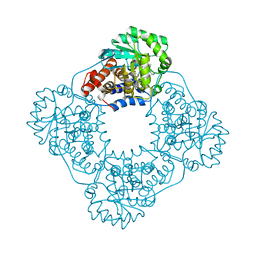 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 4 | | Descriptor: | 5-[methyl-[(2-propoxypyridin-3-yl)methyl]amino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6W45
 
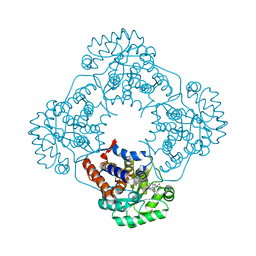 | | Crystal structure of HAO1 in complex with biaryl acid inhibitor - compound 3 | | Descriptor: | 2-chloranyl-4-[2-[[(6-chloranyl-1~{H}-indol-2-yl)carbonyl-methyl-amino]methyl]-5-fluoranyl-phenyl]benzoic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6W4C
 
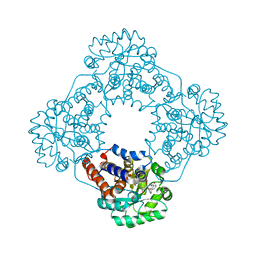 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 5 | | Descriptor: | 5-[[3-[3-(dimethylamino)-1,2,4-oxadiazol-5-yl]-2-oxidanyl-phenyl]methylamino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
3FBP
 
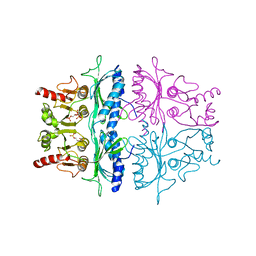 | | STRUCTURE REFINEMENT OF FRUCTOSE-1,6-BISPHOSPHATASE AND ITS FRUCTOSE 2,6-BISPHOSPHATE COMPLEX AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Ke, H, Thorpe, C.M, Seaton, B.A, Marcus, F, Lipscomb, W.N. | | Deposit date: | 1990-06-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure refinement of fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex at 2.8 A resolution.
J.Mol.Biol., 212, 1990
|
|
8IX3
 
 | | Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (local refinement) | | Descriptor: | BA.4/5 variant spike protein, heavy chain of 1G11, light chain of 1G11 | | Authors: | Sun, H, Jiang, Y, Zheng, Z, Zheng, Q, Li, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for broad neutralization of human antibody against Omicron sublineages and evasion by XBB variant.
J.Virol., 97, 2023
|
|
7QHV
 
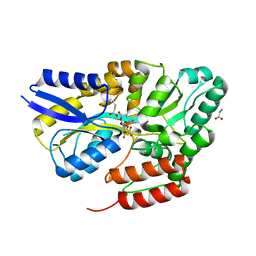 | | Crystal structure of the sulfoquinovosyl binding protein SmoF complexed with sulfoquinovosyl diacylglycerol | | Descriptor: | GLYCINE, Sulfoquinovosyl binding protein, [(2~{S},3~{S},4~{S},5~{R},6~{S})-6-[(2~{S})-3-butanoyloxy-2-heptanoyloxy-propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A, Davies, G.J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The sulfoquinovosyl glycerol binding protein SmoF binds and accommodates plant sulfolipids.
Curr Res Struct Biol, 4, 2022
|
|
7NAM
 
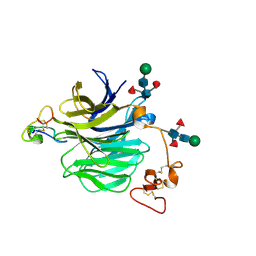 | | LRP6_E1 in complex with Lr-EET-3.5 | | Descriptor: | Low-density lipoprotein receptor-related protein 6, SODIUM ION, Trypsin inhibitor 2, ... | | Authors: | Hansen, S, Hannoush, R.N. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directed evolution identifies high-affinity cystine-knot peptide agonists and antagonists of Wnt/ beta-catenin signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6O1S
 
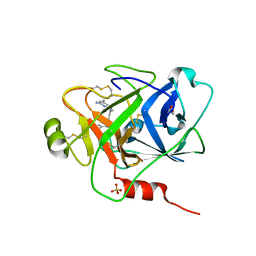 | | Structure of human plasma kallikrein protease domain with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[(6-amino-2,4-dimethylpyridin-3-yl)methyl]-1-({4-[(1H-pyrazol-1-yl)methyl]phenyl}methyl)-1H-pyrazole-4-carboxamide, PHOSPHATE ION, ... | | Authors: | Partridge, J.R, Choy, R.M. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of full-length plasma kallikrein bound to highly specific inhibitors describe a new mode of targeted inhibition.
J.Struct.Biol., 206, 2019
|
|
7M4S
 
 | |
8BNV
 
 | |
8BNX
 
 | |
8BAR
 
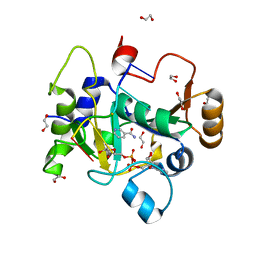 | |
8BAS
 
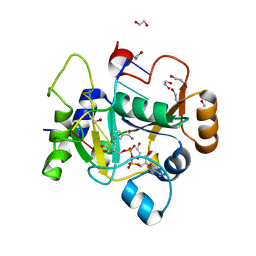 | | E. coli C7 DarT1 in complex with carba-NAD and DNA | | Descriptor: | 1,2-ETHANEDIOL, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DNA (5'-D(*AP*AP*GP*AP*C)-3'), ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
8BAT
 
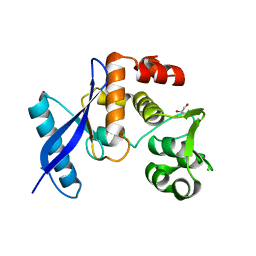 | | Geobacter lovleyi NADAR | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Geobacter lovleyi NADAR | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
8BAQ
 
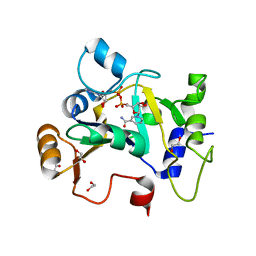 | | E. coli C7 DarT1 in complex with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, DarT ssDNA thymidine ADP-ribosyltransferase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
8BAU
 
 | | Phytophthora nicotianae var. parasitica NADAR in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, NADAR domain-containing protein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
