3MBZ
 
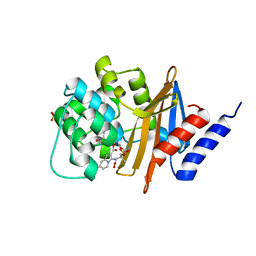 | | OXA-24 beta-lactamase complex soaked with 10mM SA4-17 inhibitor for 15min | | Descriptor: | (2S,3R)-2-[(7-aminocarbonyl-2-methanoyl-indolizin-3-yl)amino]-4-aminocarbonyloxy-3-methyl-3-sulfino-butanoic acid, Betalactamase OXA24, SULFATE ION | | Authors: | Sampson, J, van den Akker, F. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase
J.Am.Chem.Soc., 132, 2010
|
|
1HEV
 
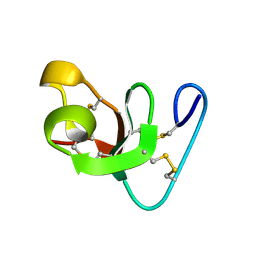 | |
3TH6
 
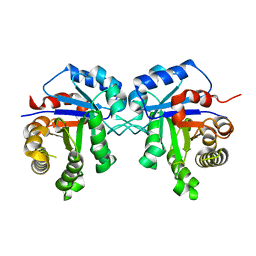 | | Crystal structure of Triosephosphate isomerase from Rhipicephalus (Boophilus) microplus. | | Descriptor: | Triosephosphate isomerase | | Authors: | Arreola, R, Rodriguez-Romero, A, Moraes, J, Gomez-Puyou, A, Perez-Montfort, R, Logullo, C. | | Deposit date: | 2011-08-18 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of a recombinant triosephosphate isomerase from Rhipicephalus (Boophilus) microplus.
Insect Biochem.Mol.Biol., 41, 2011
|
|
6OH7
 
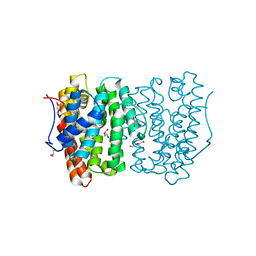 | |
3UOR
 
 | |
2ERM
 
 | | Solution structure of a biologically active human FGF-1 monomer, complexed to a hexasaccharide heparin-analogue | | Descriptor: | 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, ISOPROPYL ALCOHOL | | Authors: | Canales, A, Lozano, R, Nieto, P.M, Martin-Lomas, M, Gimenez-Gallego, G, Jimenez-Barbero, J. | | Deposit date: | 2005-10-25 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a human FGF-1 monomer, activated by a hexasaccharide heparin-analogue.
Febs J., 273, 2006
|
|
1BGG
 
 | | GLUCOSIDASE A FROM BACILLUS POLYMYXA COMPLEXED WITH GLUCONATE | | Descriptor: | BETA-GLUCOSIDASE A, D-gluconic acid | | Authors: | Sanz-Aparicio, J, Hermoso, J, Martinez-Ripoll, M, Polaina, J. | | Deposit date: | 1997-05-12 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of beta-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases.
J.Mol.Biol., 275, 1998
|
|
1BGA
 
 | |
7RAA
 
 | | Designed StabIL-2 seq15 | | Descriptor: | Interleukin-2, MAGNESIUM ION | | Authors: | Jude, K.M, Chu, A.E, Huang, P.-S, Garcia, K.C. | | Deposit date: | 2021-06-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Interleukin-2 superkines by computational design.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RA9
 
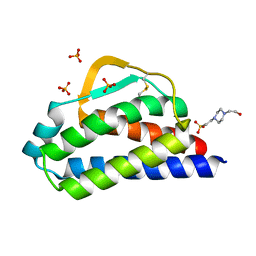 | | Designed StabIL-2 seq1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Interleukin-2, PHOSPHATE ION | | Authors: | Jude, K.M, Chu, A.E, Huang, P.-S, Garcia, K.C. | | Deposit date: | 2021-06-30 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interleukin-2 superkines by computational design.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2FT6
 
 | |
2FT8
 
 | |
2FT7
 
 | |
2FTA
 
 | |
7RDO
 
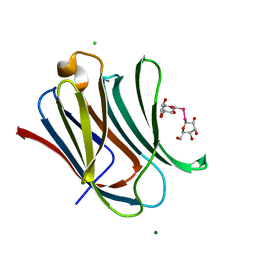 | | Crystal structure of human galectin-3 CRD in complex with diselenodigalactoside | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]diselanyl}oxane-3,4,5-triol (non-preferred name), CHLORIDE ION, Galectin-3, ... | | Authors: | Kishor, C, Go, R.M, Blanchard, H. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Investigation of the Molecular Details of the Interactions of Selenoglycosides and Human Galectin-3.
Int J Mol Sci, 23, 2022
|
|
7RDP
 
 | |
3FIM
 
 | | Crystal structure of aryl-alcohol-oxidase from Pleurotus eryingii | | Descriptor: | Aryl-alcohol oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fernandez, I.S. | | Deposit date: | 2008-12-12 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Novel structural features in the GMC family of oxidoreductases revealed by the crystal structure of fungal aryl-alcohol oxidase
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5OC1
 
 | | Crystal structure of aryl-alcohol oxidase from Pleurotus eryngii in complex with p-anisic acid | | Descriptor: | 4-METHOXYBENZOIC ACID, Aryl-alcohol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carro, J, Martinez-Julvez, M, Medina, M, Martinez, A, Ferreira, P. | | Deposit date: | 2017-06-29 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein dynamics promote hydride tunnelling in substrate oxidation by aryl-alcohol oxidase.
Phys Chem Chem Phys, 19, 2017
|
|
1XKM
 
 | | NMR structure of antimicrobial peptide distinctin in water | | Descriptor: | Distinctin chain A, Distinctin chain B | | Authors: | Amodeo, P, Raimondo, D, Andreotti, G, Motta, A, Scaloni, A. | | Deposit date: | 2004-09-29 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A folding-dependent mechanism of antimicrobial peptide resistance to degradation unveiled by solution structure of distinctin.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WRA
 
 | |
