4FBZ
 
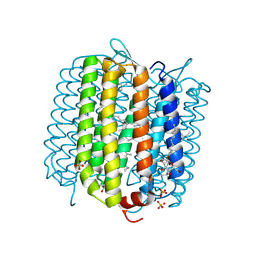 | | Crystal structure of deltarhodopsin from Haloterrigena thermotolerans | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, BACTERIORUBERIN, ... | | Authors: | Kouyama, T. | | Deposit date: | 2012-05-23 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of deltarhodopsin-3 from Haloterrigena thermotolerans
Proteins, 81, 2013
|
|
2ZZL
 
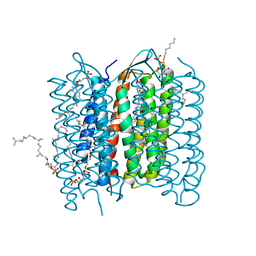 | | Structure of bacteriorhodopsin's M intermediate at pH 7 | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-O-sulfo-beta-D-galactopyranose-(1-6)-alpha-D-mannopyranose-(1-2)-alpha-D-glucopyranose, ... | | Authors: | Yamamoto, M, Kouyama, T. | | Deposit date: | 2009-02-18 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structures of Different Substates of Bacteriorhodopsin's M Intermediate at Various pH Levels.
J.Mol.Biol., 2009
|
|
3ATR
 
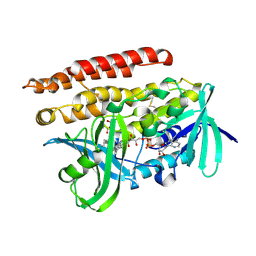 | | Geranylgeranyl Reductase (GGR) from Sulfolobus acidocaldarius co-crystallized with its ligand | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, PYROPHOSPHATE, ... | | Authors: | Fujihashi, M, Sasaki, D, Iwata, Y, Yoshimura, T, Hemmi, H, Miki, K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mutation analysis of archaeal geranylgeranyl reductase
J.Mol.Biol., 409, 2011
|
|
3AI4
 
 | | Crystal structure of yeast enhanced green fluorescent protein - mouse polymerase iota ubiquitin binding motif fusion protein | | Descriptor: | SULFATE ION, yeast enhanced green fluorescent protein,DNA polymerase iota | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AI5
 
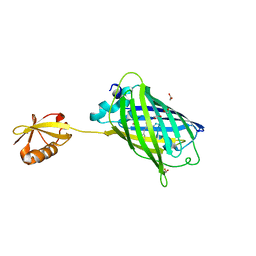 | | Crystal structure of yeast enhanced green fluorescent protein-ubiquitin fusion protein | | Descriptor: | 1,2-ETHANEDIOL, yeast enhanced green fluorescent protein,Ubiquitin | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5ETZ
 
 | |
6SGK
 
 | | Nek2 kinase bound to inhibitor 102 | | Descriptor: | 2-phenylazanyl-9~{H}-purine-6-carbonitrile, Serine/threonine-protein kinase Nek2 | | Authors: | Richards, M.W, Mas-Droux, C.P, Bayliss, R. | | Deposit date: | 2019-08-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Arylamino-6-ethynylpurines are cysteine-targeting irreversible inhibitors of Nek2 kinase.
Rsc Med Chem, 11, 2020
|
|
6SGD
 
 | | Nek2 kinase covalently bound to 2-arylamino-6-ethynylpurine inhibitor 24 | | Descriptor: | 4-[(6-ethenyl-7~{H}-purin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, SODIUM ION, ... | | Authors: | Richards, M.W, Mas-Droux, C.P, Bayliss, R. | | Deposit date: | 2019-08-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Arylamino-6-ethynylpurines are cysteine-targeting irreversible inhibitors of Nek2 kinase.
Rsc Med Chem, 11, 2020
|
|
6SGI
 
 | | Nek2 kinase bound to inhibitor 96 | | Descriptor: | 4-[(6-ethyl-7~{H}-purin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, Serine/threonine-protein kinase Nek2 | | Authors: | Richards, M.W, Mas-Droux, C.P, Bayliss, R. | | Deposit date: | 2019-08-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Arylamino-6-ethynylpurines are cysteine-targeting irreversible inhibitors of Nek2 kinase.
Rsc Med Chem, 11, 2020
|
|
6SGH
 
 | |
7DPJ
 
 | | H-Ras Q61L in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Taniguchi, H, Matsumoto, S, Miyamoto, R, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
7DPH
 
 | | H-Ras Q61H in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Taniguchi, H, Matsumoto, S, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
6FEK
 
 | |
3QBI
 
 | |
3QBL
 
 | | Pharaonis halorhodopsin complexed with nitrate | | Descriptor: | BACTERIORUBERIN, Halorhodopsin, NITRATE ION, ... | | Authors: | Kouyama, T, Kanada, S. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of anion-free blue and yellow forms of pharaonis halorhodopsin
To be Published
|
|
3QBK
 
 | | Bromide-bound form of pharaonis halorhodopsin | | Descriptor: | BACTERIORUBERIN, BROMIDE ION, Halorhodopsin, ... | | Authors: | Kouyama, T, Kanada, S. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of an O-like blue form and an anion-free yellow form of pharaonis halorhodopsin
J.Mol.Biol., 413, 2011
|
|
3QBG
 
 | | Anion-free blue form of pharaonis halorhodopsin | | Descriptor: | BACTERIORUBERIN, Halorhodopsin, RETINAL, ... | | Authors: | Kouyama, T, Kanada, S. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of an O-like blue form and an anion-free yellow form of pharaonis halorhodopsin
J.Mol.Biol., 413, 2011
|
|
5B0W
 
 | |
4L35
 
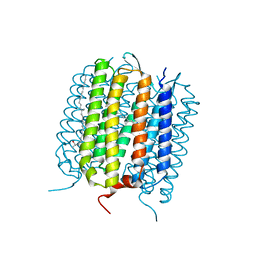 | |
4JR8
 
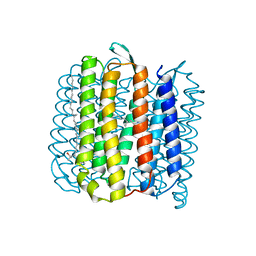 | |
1IW6
 
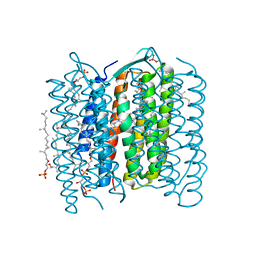 | | Crystal Structure of the Ground State of Bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Kouyama, T, Okumura, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-22 | | Release date: | 2002-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specific Damage Induced by X-ray Radiation and Structural Changes in the Primary
Photoreaction of Bacteriorhodopsin.
J.Mol.Biol., 324, 2002
|
|
1IOM
 
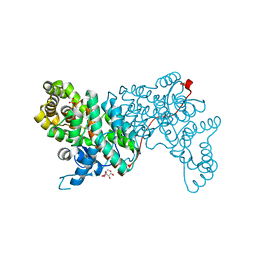 | |
2RRH
 
 | | NMR structure of vasoactive intestinal peptide in Methanol | | Descriptor: | VIP peptides | | Authors: | Umetsu, Y, Tenno, T, Goda, N, Ikegami, T, Hiroaki, H. | | Deposit date: | 2010-11-12 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural difference of vasoactive intestinal peptide in two distinct membrane-mimicking environments.
Biochim.Biophys.Acta, 1814, 2011
|
|
7JYR
 
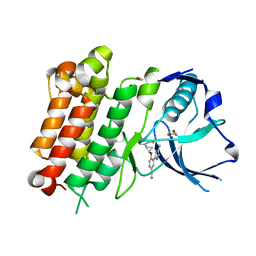 | | hALK in complex with 1-[(1R,2R)-1-(2,4-difluorophenyl)-2-[2-(5-methyl-1H-pyrazol-3-yl)-4-(trifluoromethyl)phenoxy]cyclopropyl]methanamine | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(1R,2R)-2-(2,4-difluorophenyl)cyclopropyl]oxy}-3-(5-methyl-1H-pyrazol-3-yl)benzonitrile, ALK tyrosine kinase receptor | | Authors: | McGrath, A.P, Zou, H, Lane, W, Saikatendu, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Discovery of Novel and Highly Selective Cyclopropane ALK Inhibitors through a Fragment-Assisted, Structure-Based Drug Design.
Acs Omega, 5, 2020
|
|
7JYT
 
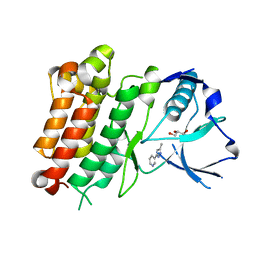 | | hALK in complex with 3-(3-methyl-1H-pyrazol-5-yl)pyridine | | Descriptor: | 3-(3-methyl-1H-pyrazol-5-yl)pyridine, ALK tyrosine kinase receptor, GLYCEROL | | Authors: | McGrath, A.P, Zou, H, Lane, W, Saikatendu, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel and Highly Selective Cyclopropane ALK Inhibitors through a Fragment-Assisted, Structure-Based Drug Design.
Acs Omega, 5, 2020
|
|
