8ETC
 
 | | Fkbp39 associated nascent 60S ribosome State 4 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUY
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUI
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 3 | | Descriptor: | 60S ribosomal protein L10-A, 60S ribosomal protein L11-A, 60S ribosomal protein L13, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
7XHM
 
 | | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with behenic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with behenic acid
To Be Published
|
|
7XHU
 
 | | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with tricosanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-04-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with tricosanoic acid
To Be Published
|
|
7WDJ
 
 | | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with linoleic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with linoleic acid
To Be Published
|
|
7WCI
 
 | | The 0.85 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with two molecules of pelargonic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Takahashi, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The 0.85 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with two molecules of pelargonic acid
To Be Published
|
|
7WC3
 
 | | X-ray structure of the human adipocyte fatty acid-binding protein complexed with the fluorescent probe HA728 | | Descriptor: | (3S)-1-(4-nitro-2,1,3-benzoxadiazol-7-yl)piperidine-3-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Takahashi, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M, Sugiyama, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of the human adipocyte fatty acid-binding protein complexed with the fluorescent probe HA728
To Be Published
|
|
7WD6
 
 | | The 0.95 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with hexanoic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXANOIC ACID, ... | | Authors: | Sugiyama, S, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The 0.95 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with hexanoic acid
To Be Published
|
|
7WE5
 
 | | The 0.87 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with oleic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with linoleic acid
To Be Published
|
|
7WF0
 
 | | The 0.83 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with nervonic acid | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-12-24 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | The 0.83 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with nervonic acid
To Be Published
|
|
7Y9Y
 
 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (no PFS) complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, RNA (27-MER), ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7Y9X
 
 | | Structure of the Cas7-11-Csx29-guide RNA complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, ZINC ION, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7CT4
 
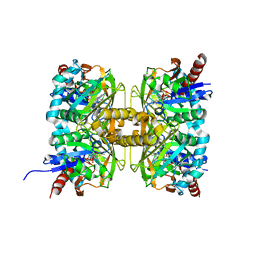 | | Crystal structure of D-amino acid oxidase from Rasamsonia emersonii strain YA | | Descriptor: | D-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Shimekake, Y, Hirato, Y, Okazaki, S, Funabashi, R, Goto, M, Furuichi, T, Suzuki, H, Takahashi, S. | | Deposit date: | 2020-08-18 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of a unique D-amino-acid oxidase from the thermophilic fungus Rasamsonia emersonii strain YA.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2RUJ
 
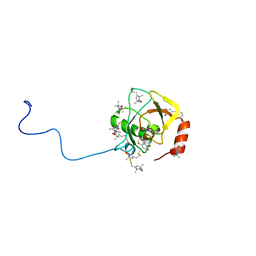 | | Solution structure of MTSL spin-labeled Schizosaccharomyces pombe Sin1 CRIM domain | | Descriptor: | Stress-activated map kinase-interacting protein 1 | | Authors: | Furuita, K, Kataoka, S, Sugiki, T, Kobayashi, N, Ikegami, T, Shiozaki, K, Fujiwara, T, Kojima, C. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Method: | SOLUTION NMR | | Cite: | Utilization of paramagnetic relaxation enhancements for high-resolution NMR structure determination of a soluble loop-rich protein with sparse NOE distance restraints
J.Biomol.Nmr, 61, 2015
|
|
2D7I
 
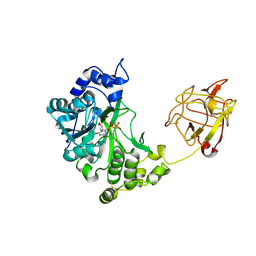 | | Crystal structure of pp-GalNAc-T10 with UDP, GalNAc and Mn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kubota, T, Shiba, T, Sugioka, S, Kato, R, Wakatsuki, S, Narimatsu, H. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10)
J.Mol.Biol., 359, 2006
|
|
2D7R
 
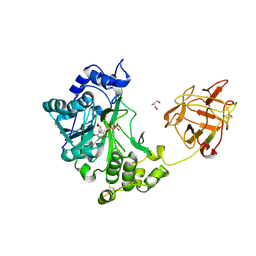 | | Crystal structure of pp-GalNAc-T10 complexed with GalNAc-Ser on lectin domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kubota, T, Shiba, T, Sugioka, S, Kato, R, Wakatsuki, S, Narimatsu, H. | | Deposit date: | 2005-11-25 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10)
J.Mol.Biol., 359, 2006
|
|
4I9A
 
 | | Crystal Structure of Sus scrofa Quinolinate Phosphoribosyltransferase in Complex with Nicotinate Mononucleotide | | Descriptor: | NICOTINATE MONONUCLEOTIDE, quinolinate phosphoribosyltransferase | | Authors: | Youn, H.-S, Kim, M.-K, Kang, K.B, Kim, T.G, Lee, J.-G, An, J.Y, Park, K.R, Lee, Y, Kang, J.Y, Song, H.E, Park, I, Cho, C, Fukuoka, S, Eom, S.H. | | Deposit date: | 2012-12-05 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Crystal structure of Sus scrofa quinolinate phosphoribosyltransferase in complex with nicotinate mononucleotide
Plos One, 8, 2013
|
|
5ZBD
 
 | | Crystal structure of tryptophan oxidase (C395A mutant) from Chromobacterium violaceum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, TRYPTOPHAN | | Authors: | Yamaguchi, H, Tatsumi, M, Takahashi, K, Tagami, U, Sugiki, M, Kashiwagi, T, Okazaki, S, Mizukoshi, T, Asano, Y. | | Deposit date: | 2018-02-11 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein engineering for improving the thermostability of tryptophan oxidase and insights from structural analysis.
J. Biochem., 164, 2018
|
|
5ZBC
 
 | | Crystal structure of Se-Met tryptophan oxidase (C395A mutant) from Chromobacterium violaceum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA | | Authors: | Yamaguchi, H, Tatsumi, M, Takahashi, K, Tagami, U, Sugiki, M, Kashiwagi, T, Okazaki, S, Mizukoshi, T, Asano, Y. | | Deposit date: | 2018-02-11 | | Release date: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein engineering for improving the thermostability of tryptophan oxidase and insights from structural analysis.
J. Biochem., 164, 2018
|
|
1IOO
 
 | | CRYSTAL STRUCTURE OF NICOTIANA ALATA GEMETOPHYTIC SELF-INCOMPATIBILITY ASSOCIATED SF11-RNASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-xylopyranose-(1-2)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SF11-RNASE, beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ida, K, Sato, M, Sakiyama, F, Norioka, S, Yamamoto, M, Kumasaka, T, Yamashita, E. | | Deposit date: | 2001-03-26 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The 1.55 A resolution structure of Nicotiana alata S(F11)-RNase associated with gametophytic self-incompatibility.
J.Mol.Biol., 314, 2001
|
|
1IQQ
 
 | | Crystal Structure of Japanese pear S3-RNase | | Descriptor: | S3-RNase, beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Matsuura, T, Sakai, H, Norioka, S. | | Deposit date: | 2001-07-25 | | Release date: | 2001-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure at 1.5-A resolution of Pyrus pyrifolia pistil ribonuclease responsible for gametophytic self-incompatibility.
J.Biol.Chem., 276, 2001
|
|
1FZZ
 
 | | THE CRYSTAL STRUCTURE OF THE COMPLEX OF NON-PEPTIDIC INHIBITOR ONO-6818 AND PORCINE PANCREATIC ELASTASE. | | Descriptor: | 2-(5-AMINO-6-OXO-2-PHENYL-6H-PYRIMIDIN-1-YL)-N-[2-(5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)-1-(METHYLETHYL)-2-HYDROXYETHYL]ACETAMIDE, ELASTASE 1 | | Authors: | Odagaki, Y, Ohmoto, K, Matsuoka, S, Hamanaka, N, Nakai, H, Toda, M, Katsuya, Y. | | Deposit date: | 2000-10-04 | | Release date: | 2001-10-04 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The crystal structure of the complex of non-peptidic inhibitor of human neutrophil elastase ONO-6818 and porcine pancreatic elastase.
Bioorg.Med.Chem., 9, 2001
|
|
3WMW
 
 | | GalE-like L-Threonine dehydrogenase from Cupriavidus necator (apo form) | | Descriptor: | CALCIUM ION, NAD dependent epimerase/dehydratase | | Authors: | Nakano, S, Okazaki, S, Tokiwa, H, Asano, Y. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding of NAD+ and L-Threonine Induces Stepwise Structural and Flexibility Changes in Cupriavidus necator L-Threonine Dehydrogenase
J.Biol.Chem., 289, 2014
|
|
3WMX
 
 | | GalE-like L-Threonine dehydrogenase from Cupriavidus necator (holo form) | | Descriptor: | NAD dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, THREONINE | | Authors: | Nakano, S, Okazaki, S, Tokiwa, H, Asano, Y. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding of NAD+ and L-Threonine Induces Stepwise Structural and Flexibility Changes in Cupriavidus necator L-Threonine Dehydrogenase
J.Biol.Chem., 289, 2014
|
|
