1V4X
 
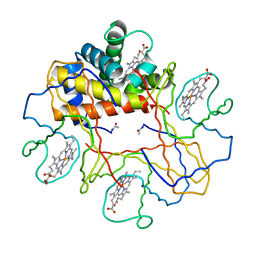 | | Crystal structure of bluefin tuna hemoglobin deoxy form at pH5.0 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha chain, hemoglobin beta chain | | Authors: | Yokoyama, T, Chong, K.T, Miyazaki, Y, Nakatsukasa, T, Unzai, S, Miyazaki, G, Morimoto, H, Jeremy, R.H.T, Park, S.Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel Mechanisms of pH Sensitivity in Tuna Hemoglobin: A STRUCTURAL EXPLANATION OF THE ROOT EFFECT
J.Biol.Chem., 279, 2004
|
|
1V4U
 
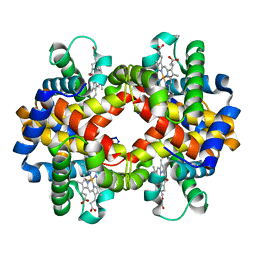 | | Crystal structure of bluefin tuna carbonmonoxy-hemoglobin | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha chain, ... | | Authors: | Yokoyama, T, Chong, K.T, Miyazaki, Y, Nakatsukasa, T, Unzai, S, Miyazaki, G, Morimoto, H, Jeremy, R.H.T, Park, S.Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Mechanisms of pH Sensitivity in Tuna Hemoglobin: A STRUCTURAL EXPLANATION OF THE ROOT EFFECT
J.Biol.Chem., 279, 2004
|
|
3C3I
 
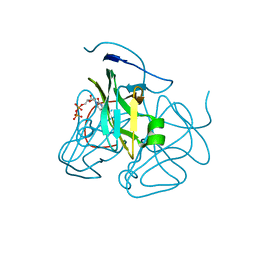 | | Evolution of chlorella virus dUTPase | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | Authors: | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | Deposit date: | 2008-01-28 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5AUS
 
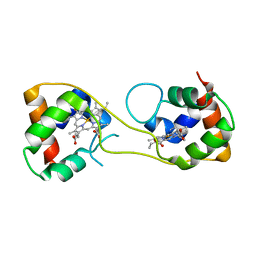 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at C-terminal region | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
2ZVM
 
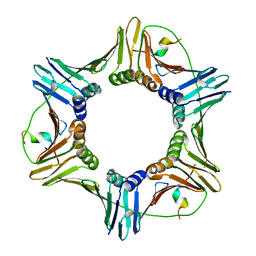 | | Crystal structure of PCNA in complex with DNA polymerase iota fragment | | Descriptor: | DNA polymerase iota, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
2ZVK
 
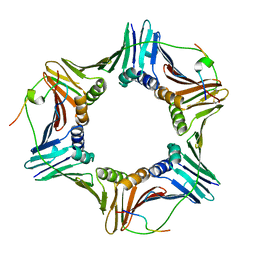 | | Crystal structure of PCNA in complex with DNA polymerase eta fragment | | Descriptor: | DNA polymerase eta, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
1VDZ
 
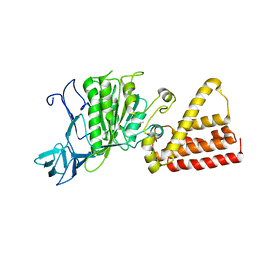 | | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, A-type ATPase subunit A | | Authors: | Maegawa, Y, Morita, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3
To be Published
|
|
7V5Q
 
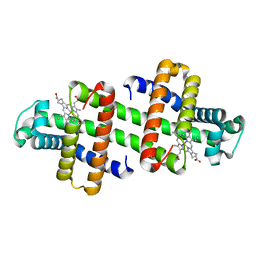 | | The dimeric structure of G80A/H81A/L137E myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xie, C, Komori, H, Hirota, S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Experimental and theoretical study on converting myoglobin into a stable domain-swapped dimer by utilizing a tight hydrogen bond network at the hinge region.
Rsc Adv, 11, 2021
|
|
7V5R
 
 | | The dimeric structure of G80A/H81A/L137D myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xie, C, Komori, H, Hirota, S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Experimental and theoretical study on converting myoglobin into a stable domain-swapped dimer by utilizing a tight hydrogen bond network at the hinge region.
Rsc Adv, 11, 2021
|
|
1J1J
 
 | | Crystal Structure of human Translin | | Descriptor: | Translin | | Authors: | Sugiura, I, Sasaki, C, Hasegawa, T, Kohno, T, Sugio, S, Moriyama, H, Kasai, M, Matsuzaki, T. | | Deposit date: | 2002-12-06 | | Release date: | 2003-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human translin at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1IZR
 
 | | F46A mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
1IZP
 
 | | F46L mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
1IDM
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE, LOOP-DELETED CHIMERA | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Sakurai, M, Ohzeki, M, Moriyama, H, Sato, M, Tanaka, N. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a loop-deleted variant of 3-isopropylmalate dehydrogenase from Thermus thermophilus: an internal reprieve tolerance mechanism.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1JS1
 
 | | Crystal Structure of a new transcarbamylase from the anaerobic bacterium Bacteroides fragilis at 2.0 A resolution | | Descriptor: | PHOSPHATE ION, Transcarbamylase | | Authors: | Shi, D, Gallegos, R, DePonte III, J, Morizono, H, Yu, X, Allewell, N.M, Malamy, M, Tuchman, M. | | Deposit date: | 2001-08-15 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a transcarbamylase-like protein from the anaerobic bacterium Bacteroides fragilis at 2.0 A resolution.
J.Mol.Biol., 320, 2002
|
|
1XER
 
 | | STRUCTURE OF FERREDOXIN | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, ZINC ION | | Authors: | Fujii, T, Hata, Y, Moriyama, H, Wakagi, T, Tanaka, N, Oshima, T. | | Deposit date: | 1996-08-28 | | Release date: | 1997-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding centre in thermoacidophilic archaeal ferredoxins.
Nat.Struct.Biol., 3, 1996
|
|
1IZQ
 
 | | F46V mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
6ICH
 
 | | Grb2 SH2 domain in domain swapped dimer form | | Descriptor: | Growth factor receptor-bound protein 2 | | Authors: | Hosoe, Y, Numoto, N, Inaba, S, Ogawa, S, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2018-09-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional properties of Grb2 SH2 dimer in CD28 binding.
Biophys Physicobio., 16, 2019
|
|
6ICG
 
 | | Grb2 SH2 domain in phosphopeptide free form | | Descriptor: | GLYCEROL, Growth factor receptor-bound protein 2, SULFATE ION | | Authors: | Hosoe, Y, Numoto, N, Inaba, S, Ogawa, S, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2018-09-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and functional properties of Grb2 SH2 dimer in CD28 binding.
Biophys Physicobio., 16, 2019
|
|
6LFX
 
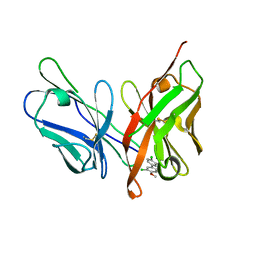 | |
3VKP
 
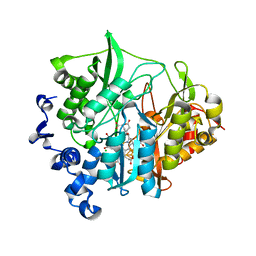 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with low X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
3X15
 
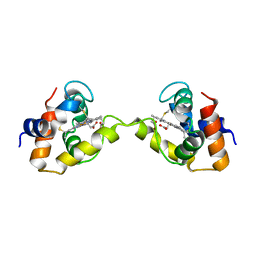 | | Dimeric Aquifex aeolicus cytochrome c555 | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Yamanaka, M, Nagao, S, Komori, H, Higuchi, Y, Hirota, S. | | Deposit date: | 2014-10-29 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Change in structure and ligand binding properties of hyperstable cytochrome c555 from Aquifex aeolicus by domain swapping
Protein Sci., 24, 2015
|
|
2DT5
 
 | | Crystal Structure of TTHA1657 (AT-rich DNA-binding protein) from Thermus thermophilus HB8 | | Descriptor: | AT-rich DNA-binding protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nakamura, A, Sosa, A, Komori, H, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-11 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of TTHA1657 (AT-rich DNA-binding protein; p25) from Thermus thermophilus HB8 at 2.16 A resolution
Proteins, 66, 2007
|
|
2YXW
 
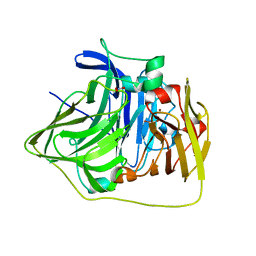 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
7W6F
 
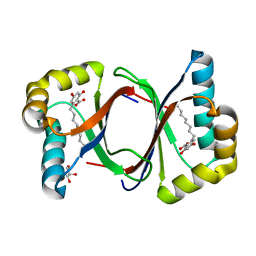 | | Polyketide cyclase OAC-F24I mutant from Cannabis sativa in complex with 6-nonylresorcylic acid | | Descriptor: | 2-nonyl-4,6-bis(oxidanyl)benzoic acid, GLYCEROL, Olivetolic acid cyclase | | Authors: | Nakashima, Y, Lee, Y.E, Morita, H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual Engineering of Olivetolic Acid Cyclase and Tetraketide Synthase to Generate Longer Alkyl-Chain Olivetolic Acid Analogs.
Org.Lett., 24, 2022
|
|
7W6G
 
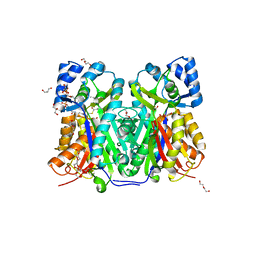 | | TKS-L190G mutant from Cannabis sativa in complex with lauroyl-CoA | | Descriptor: | 3,5,7-trioxododecanoyl-CoA synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakashima, Y, Lee, Y.E, Morita, H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Engineering of Olivetolic Acid Cyclase and Tetraketide Synthase to Generate Longer Alkyl-Chain Olivetolic Acid Analogs.
Org.Lett., 24, 2022
|
|
