3WHC
 
 | |
3WQP
 
 | | Crystal structure of Rubisco T289D mutant from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujihashi, M, Nishitani, Y, Kiriyama, T, Miki, K. | | Deposit date: | 2014-01-29 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mutation design of thermophilic Rubisco based on the three-dimensional structure enhances its activity at ambient temperature
to be published
|
|
3WHB
 
 | |
3W4S
 
 | |
3A12
 
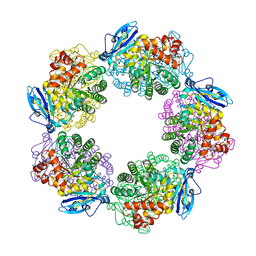 | | Crystal structure of Type III Rubisco complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
2ZDI
 
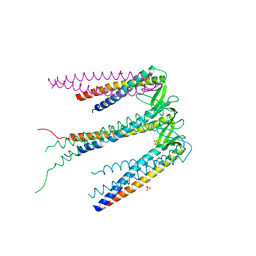 | | Crystal structure of Prefoldin from Pyrococcus horikoshii OT3 | | Descriptor: | Prefoldin subunit alpha, Prefoldin subunit beta, SULFATE ION | | Authors: | Kida, H, Miki, K. | | Deposit date: | 2007-11-23 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and molecular dynamics simulation of archaeal prefoldin: the molecular mechanism for binding and recognition of nonnative substrate proteins
J.Mol.Biol., 376, 2008
|
|
2Z9O
 
 | |
3VM6
 
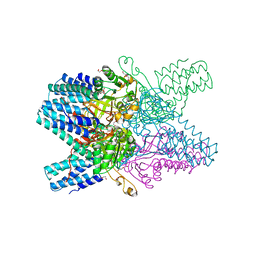 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1 in complex with alpha-D-ribose-1,5-bisphosphate | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, A, Fujihashi, M, Aono, R, Sato, T, Nishiba, Y, Yoshida, S, Yano, A, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2011-12-08 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1
J.Biol.Chem., 287, 2012
|
|
2ZFO
 
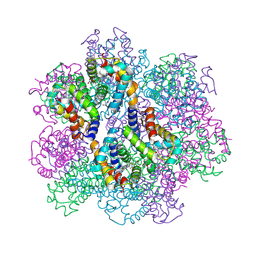 | | Structure of the partially unliganded met state of 400 kDa hemoglobin: Insights into ligand-induced structural changes of giant hemoglobins | | Descriptor: | Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, Extracellular giant hemoglobin major globin subunit B1, ... | | Authors: | Numoto, N, Nakagawa, T, Kita, A, Sasayama, Y, Fukumori, Y, Miki, K. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the partially unliganded met state of 400 kDa hemoglobin: insights into ligand-induced structural changes of giant hemoglobins
Proteins, 73, 2008
|
|
3VYT
 
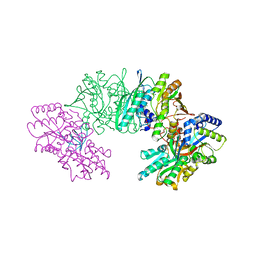 | | Crystal structure of the HypC-HypD-HypE complex (form I inward) | | Descriptor: | CHLORIDE ION, Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3VRC
 
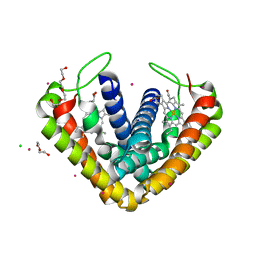 | | Crystal structure of cytochrome c' from Thermochromatium tepidum | | Descriptor: | CADMIUM ION, CHLORIDE ION, Cytochrome c', ... | | Authors: | Hirano, Y, Kimura, Y, Suzuki, H, Miki, K, Wang, Z.-Y. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Analysis and Comparative Characterization of the Cytochrome c' and Flavocytochrome c from Thermophilic Purple Photosynthetic Bacterium Thermochromatium tepidum
Biochemistry, 51, 2012
|
|
3VYR
 
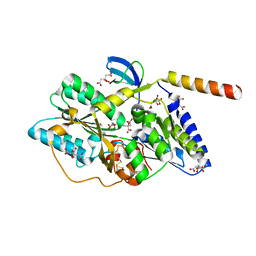 | | Crystal structure of the HypC-HypD complex | | Descriptor: | CITRIC ACID, Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3VRD
 
 | | Crystal structure of flavocytochrome c from Thermochromatium tepidum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavocytochrome c flavin subunit, Flavocytochrome c heme subunit, ... | | Authors: | Hirano, Y, Kimura, Y, Suzuki, H, Miki, K, Wang, Z.-Y. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure Analysis and Comparative Characterization of the Cytochrome c' and Flavocytochrome c from Thermophilic Purple Photosynthetic Bacterium Thermochromatium tepidum
Biochemistry, 51, 2012
|
|
3VYS
 
 | | Crystal structure of the HypC-HypD-HypE complex (form I) | | Descriptor: | Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, Hydrogenase expression/formation protein HypE, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3A11
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Thermococcus kodakaraensis KOD1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Translation initiation factor eIF-2B, ... | | Authors: | Nakamura, A, Fujihashi, M, Nishiba, Y, Yoshida, S, Yano, A, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-03-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1
J.Biol.Chem., 287, 2012
|
|
3VYU
 
 | | Crystal structure of the HypC-HypD-HypE complex (form II) | | Descriptor: | Hydrogenase expression/formation protein HypC, Hydrogenase expression/formation protein HypD, Hydrogenase expression/formation protein HypE, ... | | Authors: | Watanabe, S, Miki, K. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation
Structure, 20, 2012
|
|
3A43
 
 | | Crystal structure of HypA | | Descriptor: | Hydrogenase nickel incorporation protein hypA, ZINC ION | | Authors: | Watanabe, S, Arai, T, Matsumi, R, Aromi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-06-30 | | Release date: | 2009-10-06 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation.
J.Mol.Biol., 394, 2009
|
|
3WDK
 
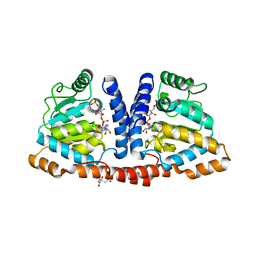 | | Crystal structure of 4-phosphopantoate-beta-alanine ligase complexed with reaction intermediate | | Descriptor: | 4-phosphopantoate--beta-alanine ligase, 5'-O-[(S)-hydroxy{[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]oxy}phosphoryl]adenosine, CITRATE ANION | | Authors: | Kishimoto, A, Kita, A, Ishibashi, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis
Proteins, 82, 2014
|
|
3WJP
 
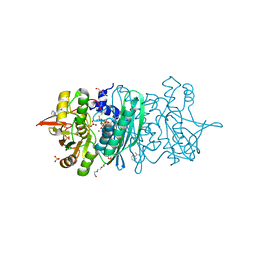 | | Crystal structure of the HypE CA form | | Descriptor: | BENZAMIDINE, GLYCEROL, Hydrogenase expression/formation protein HypE, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WK1
 
 | |
3WJR
 
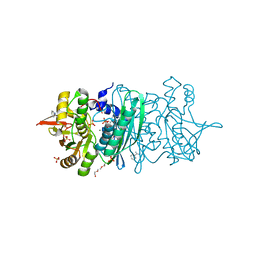 | | crystal structure of HypE in complex with a nucleotide | | Descriptor: | ADENOSINE MONOPHOSPHATE, BENZAMIDINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WJZ
 
 | | Orotidine 5'-monophosphate decarboxylase D75N mutant from M. thermoautotrophicus complexed with 6-amino-UMP | | Descriptor: | 6-AMINOURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fujihashi, M, Pai, E.F, Miki, K. | | Deposit date: | 2013-10-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate distortion contributes to the catalysis of orotidine 5'-monophosphate decarboxylase.
J.Am.Chem.Soc., 135, 2013
|
|
3A13
 
 | | Crystal structure of Type III Rubisco SP4 mutant complexed with 2-CABP and activated with Ca | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based optimization of a Type III Rubisco from a hyperthermophile
To be Published
|
|
3A9F
 
 | | Crystal structure of the C-terminal domain of cytochrome cz from Chlorobium tepidum | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Cytochrome c, HEME C, ... | | Authors: | Hirano, Y, Higuchi, M, Azai, C, Oh-oka, H, Miki, K, Wang, Z.-Y. | | Deposit date: | 2009-10-25 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the electron carrier domain of the reaction center cytochrome c(z) subunit from green photosynthetic bacterium Chlorobium tepidum
J.Mol.Biol., 397, 2010
|
|
3WDM
 
 | | Crystal structure of 4-phosphopantoate-beta-alanine ligase from Thermococcus kodakarensis | | Descriptor: | 4-phosphopantoate--beta-alanine ligase, ADENOSINE | | Authors: | Kishimoto, A, Kita, A, Ishibashi, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis
Proteins, 82, 2014
|
|
