3K13
 
 | | Structure of the pterin-binding domain MeTr of 5-methyltetrahydrofolate-homocysteine methyltransferase from Bacteroides thetaiotaomicron | | Descriptor: | 5-methyltetrahydrofolate-homocysteine methyltransferase, GLYCEROL, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, ... | | Authors: | Cuff, M.E, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-25 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the pterin-binding domain MeTr of 5-methyltetrahydrofolate-homocysteine methyltransferase from Bacteroides thetaiotaomicron
TO BE PUBLISHED
|
|
3CRJ
 
 | |
3DOB
 
 | | Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans. | | Descriptor: | BETA-MERCAPTOETHANOL, Heat shock 70 kDa protein F44E5.5 | | Authors: | Osipiuk, J, Hatzos, C, Gu, M, Zhang, R, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-03 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray crystal structure of Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans.
To be Published
|
|
3K67
 
 | | Crystal structure of protein af1124 from archaeoglobus fulgidus | | Descriptor: | PHOSPHATE ION, putative dehydratase AF1124 | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Protein Af1124 from Archaeoglobus Fulgidus
To be Published
|
|
3D3R
 
 | | Crystal structure of the hydrogenase assembly chaperone HypC/HupF family protein from Shewanella oneidensis MR-1 | | Descriptor: | Hydrogenase assembly chaperone hypC/hupF | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Hydrogenase Assembly Chaperone HypC/HupF Family Protein from Shewanella oneidensis MR-1.
To be Published
|
|
3KAO
 
 | | Crystal structure of tagatose 1,6-diphosphate aldolase from Staphylococcus aureus | | Descriptor: | GLYCEROL, SULFATE ION, Tagatose 1,6-diphosphate aldolase, ... | | Authors: | Chang, C, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of tagatose 1,6-diphosphate aldolase from Staphylococcus aureus
To be Published
|
|
3K1U
 
 | | Beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-xylosidase, ... | | Authors: | Osipiuk, J, Wu, R, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of
beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum at 1.55 A resolution
To be Published
|
|
3K3S
 
 | | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri. | | Descriptor: | ACETATE ION, Altronate hydrolase, CHLORIDE ION, ... | | Authors: | Hou, J, Chruszcz, M, Xu, X, Le, B, Zimmerman, M.D, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri.
To be Published
|
|
1KXJ
 
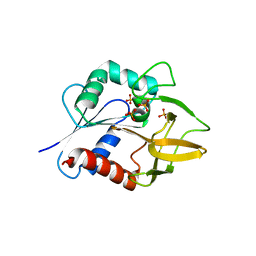 | | The Crystal Structure of Glutamine Amidotransferase from Thermotoga maritima | | Descriptor: | Amidotransferase hisH, PHOSPHATE ION | | Authors: | Korolev, S, Skarina, T, Evdokimova, E, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-31 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glutamine amidotransferase from Thermotoga maritima
Proteins, 49, 2002
|
|
3D7N
 
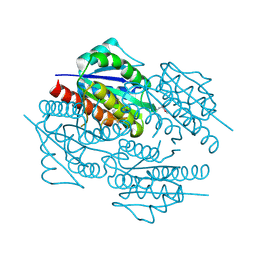 | | The crystal structure of the flavodoxin, WrbA-like protein from Agrobacterium tumefaciens | | Descriptor: | Flavodoxin, WrbA-like protein | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-21 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the flavodoxin, WrbA-like protein from Agrobacterium tumefaciens
To be Published, 2008
|
|
3SFP
 
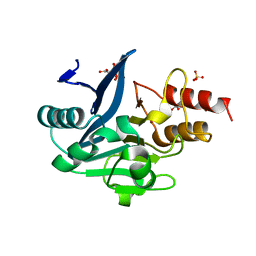 | | Crystal Structure of the Mono-Zinc-boundform of New Delhi Metallo-beta-Lactamase-1 from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-06-13 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
1KJN
 
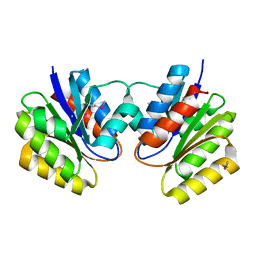 | |
3S9X
 
 | | High resolution crystal structure of ASCH domain from Lactobacillus crispatus JV V101 | | Descriptor: | ASCH domain, CHLORIDE ION | | Authors: | Nocek, B, Xu, X, Cui, H, Jedrzejczak, R, Edwards, A, Savchenko, A, Mabbutt, B.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-06-02 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution crystal structure of ASCH domain from Lactobacillus crispatus JV V101
TO BE PUBLISHED
|
|
3JSA
 
 | | Homoserine dehydrogenase from Thermoplasma volcanium complexed with NAD | | Descriptor: | Homoserine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Osipiuk, J, Nocek, B, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-09 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of Homoserine dehydrogenase from Thermoplasma volcanium
To be Published
|
|
3SJR
 
 | |
3K2N
 
 | |
3SBL
 
 | | Crystal Structure of New Delhi Metal-beta-lactamase-1 from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, CITRIC ACID | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-06-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
3S4L
 
 | | The CRISPR-associated Cas3 HD domain protein MJ0384 from Methanocaldococcus jannaschii | | Descriptor: | CALCIUM ION, CAS3 Metal dependent phosphohydrolase | | Authors: | Petit, P, Brown, G, Yakunin, A, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-05-19 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference.
Embo J., 30, 2011
|
|
4W9T
 
 | | Crystal structure of HisAP from Streptomyces sp. Mg1 | | Descriptor: | Phosphoribosyl isomerase A, SULFATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
1U7N
 
 | | Crystal Structure of the fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Fatty acid/phospholipid synthesis protein plsX | | Authors: | Kim, Y, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-04 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of fatty acid/phospholipid synthesis protein PlsX from Enterococcus faecalis.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
1TU9
 
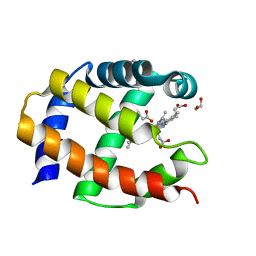 | | Crystal Structure of a Protein PA3967, a Structurally Highly Homologous to a Human Hemoglobin, from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, PROPANOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Egorova, O, Bochkarev, A, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of PA3967 from Pseudomonas aeruginosa PAO1, a Hypothetical Protein which is highly homologous to human Hemoglobin in structure.
To be Published
|
|
1U9C
 
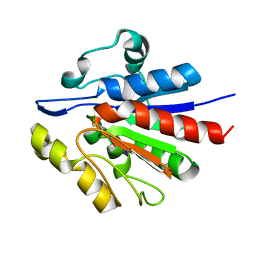 | | Crystallographic structure of APC35852 | | Descriptor: | APC35852 | | Authors: | Borek, D, Chen, Y, Shao, D, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of DJI superfamily
To be Published
|
|
1TZ0
 
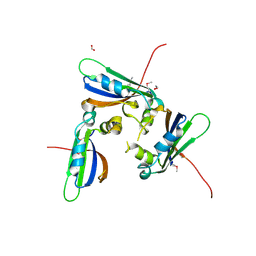 | |
1U9D
 
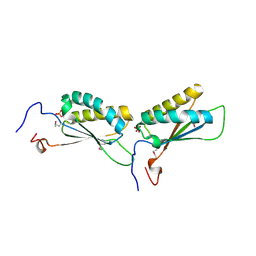 | |
1U0K
 
 | | The structure of a Predicted Epimerase PA4716 from Pseudomonas aeruginosa | | Descriptor: | gene product PA4716 | | Authors: | Cuff, M.E, Ginell, S.L, Rotella, F.J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hypothetical protein PA4716 from Pseudomonas aeruginosa
TO BE PUBLISHED
|
|
