8A74
 
 | | PcIDS1_F315A in complex with Mg2+ and GPP | | Descriptor: | GERANYL DIPHOSPHATE, Isoprenyl diphosphate synthase, MAGNESIUM ION | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
1NZC
 
 | | The high resolution structures of RmlC from Streptococcus suis in complex with dTDP-D-xylose | | Descriptor: | NICKEL (II) ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | Authors: | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | Deposit date: | 2003-02-17 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
8AID
 
 | |
5B25
 
 | | Crystal structure of human PDE1B with inhibitor 3 | | Descriptor: | (11R,15S)-4-{[4-(6-fluoropyridin-2-yl)phenyl]methyl}-8-methyl-5-(phenylamino)-1,3,4,8,10-pentaazatetracyclo[7.6.0.02,6.011,15]pentadeca-2,5,9-trien-7-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, GLYCEROL, ... | | Authors: | Ida, K, Lane, W, Snell, G, Sogabe, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Phosphodiesterase 1 for the Treatment of Cognitive Impairment Associated with Neurodegenerative and Neuropsychiatric Diseases
J.Med.Chem., 59, 2016
|
|
8AJQ
 
 | |
3NKV
 
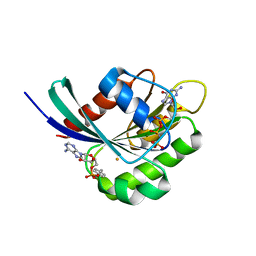 | | Crystal structure of Rab1b covalently modified with AMP at Y77 | | Descriptor: | ADENOSINE MONOPHOSPHATE, BARIUM ION, MAGNESIUM ION, ... | | Authors: | Mueller, M.P, Peters, H, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b.
Science, 329, 2010
|
|
2YJJ
 
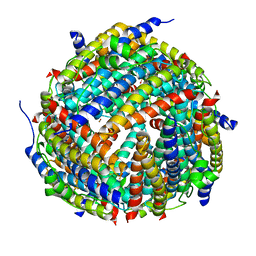 | |
2YJK
 
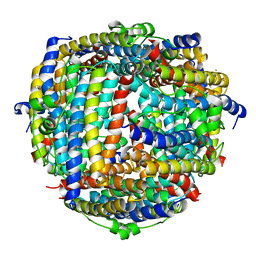 | |
1OWD
 
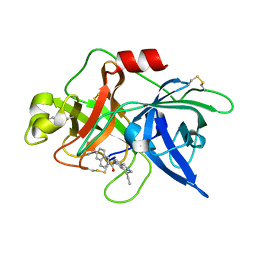 | | Substituted 2-Naphthamidine inhibitors of urokinase | | Descriptor: | 6-[AMINO(IMINO)METHYL]-N-[(4R)-4-ETHYL-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL]-2-NAPHTHAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
3NKU
 
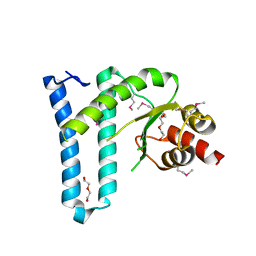 | | Crystal structure of the N-terminal domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, DrrA, TRIETHYLENE GLYCOL | | Authors: | Mueller, M.P, Peters, H, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b.
Science, 329, 2010
|
|
1NYW
 
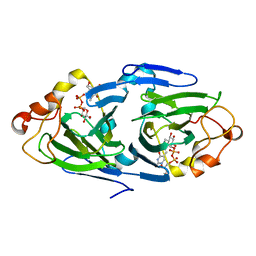 | | The high resolution structures of RmlC from Streptoccus suis in complex with dTDP-D-glucose | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | Authors: | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | Deposit date: | 2003-02-14 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
3N6O
 
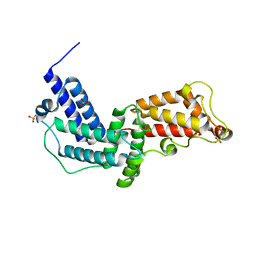 | | Crystal structure of the GEF and P4M domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | SULFATE ION, guanine nucleotide exchange factor | | Authors: | Schoebel, S, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA.
Embo Rep., 11, 2010
|
|
3KH4
 
 | |
1OWH
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-[4-(AMINOMETHYL)PHENYL]-2-NAPHTHAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
1OWE
 
 | | Substituted 2-Naphthamidine inhibitors of urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-PHENYL-2-NAPHTHAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
1OWJ
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-(1-ISOPROPYL-3,4-DIHYDROISOQUINOLIN-7-YL)-2-NAPHTHAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
1OWK
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-(1-ISOPROPYL-1,2,3,4-TETRAHYDROISOQUINOLIN-7-YL)-2-NAPHTHAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
1OWI
 
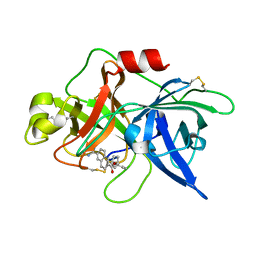 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-[3-(CYCLOPENTYLOXY)PHENYL]-2-NAPHTHAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
2Q0J
 
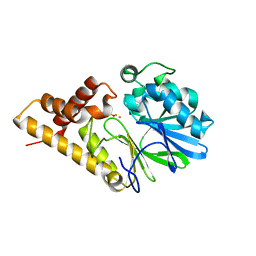 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | Descriptor: | BENZOIC ACID, FE (III) ION, Quinolone signal response protein | | Authors: | Yu, S, Jensen, V, Feldmann, I, Haussler, S, Blankenfeldt, W. | | Deposit date: | 2007-05-22 | | Release date: | 2008-06-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
5MBR
 
 | |
5MCR
 
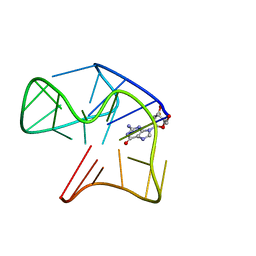 | | Quadruplex with flipped tetrad formed by an artificial sequence | | Descriptor: | Artificial quadruplex with propeller, diagonal and lateral loop | | Authors: | Dickerhoff, J, Haase, L, Langel, W, Weisz, K. | | Deposit date: | 2016-11-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Tracing Effects of Fluorine Substitutions on G-Quadruplex Conformational Changes.
ACS Chem. Biol., 12, 2017
|
|
5MZW
 
 | | Crystal structure of the decarboxylase AibA/AibB | | Descriptor: | GLYCEROL, Glutaconate CoA-transferase family, subunit A, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MZY
 
 | | Crystal structure of the decarboxylase AibA/AibB in complex with a possible transition state analog | | Descriptor: | (1~{R},2~{S})-2-methylcyclohexane-1-carboxylic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5N00
 
 | | Crystal structure of the decarboxylase AibA/AibB C56A variant | | Descriptor: | ACETATE ION, Glutaconate CoA-transferase family, subunit A, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NCH
 
 | | GriE apo form | | Descriptor: | Leucine hydroxylase | | Authors: | Lukat, P, Blankenfeldt, W, Mueller, R. | | Deposit date: | 2017-03-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity.
Chem Sci, 8, 2017
|
|
