4R2G
 
 | | Crystal Structure of PGT124 Fab bound to HIV-1 JRCSF gp120 core and to CD4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Garces, F, Wilson, I.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
5YCP
 
 | | Human PPARgamma ligand binding domain complexed with Rosiglitazone | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Nuclear receptor coactivator 1, ... | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPAR gamma.
Sci Rep, 8, 2018
|
|
6A0H
 
 | |
6A0F
 
 | |
6A0I
 
 | |
6A0E
 
 | |
5YCN
 
 | | Human PPARgamma ligand binding domain complexed with Lobeglitazone | | Descriptor: | (5S)-5-[[4-[2-[[6-(4-methoxyphenoxy)pyrimidin-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPAR gamma.
Sci Rep, 8, 2018
|
|
2XJI
 
 | | Structure and Copper-binding Properties of Methanobactins from Methylosinus trichosporium OB3b | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB-OB3B | | Authors: | El-Ghazouani, A, Basle, A, Firbank, S.J, Knapp, C.W, Gray, J, Graham, D.W, Dennison, C. | | Deposit date: | 2010-07-06 | | Release date: | 2011-02-02 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
Inorg.Chem., 50, 2011
|
|
2XJH
 
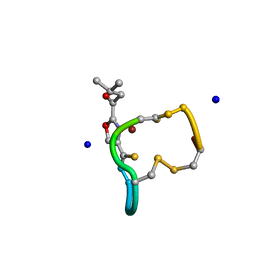 | | Structure and Copper-binding Properties of Methanobactins from Methylosinus trichosporium OB3b | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB-OB3B, SODIUM ION | | Authors: | El-Ghazouani, A, Basle, A, Firbank, S.J, Knapp, C.W, Gray, J, Graham, D.W, Dennison, C. | | Deposit date: | 2010-07-06 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
Inorg.Chem., 50, 2011
|
|
5XGW
 
 | |
5Z8A
 
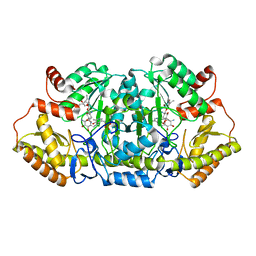 | | Crystal structure of GenB1 from Micromonospora echinospora in complex with JI-20A and PLP (external aldimine) | | Descriptor: | (2~{R},3~{R},4~{R},5~{R})-2-[(1~{S},2~{S},3~{R},4~{S},6~{R})-3-[(2~{R},3~{R},4~{R},5~{S},6~{R})-6-(aminomethyl)-3-azany l-4,5-bis(oxidanyl)oxan-2-yl]oxy-4,6-bis(azanyl)-2-oxidanyl-cyclohexyl]oxy-5-methyl-4-(methylamino)oxane-3,5-diol, C-6' aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hong, S.K, Cha, S.S. | | Deposit date: | 2018-01-31 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Complete reconstitution of the diverse pathways of gentamicin B biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
5Z2E
 
 | |
5Z2F
 
 | | NADPH/PDA bound Dihydrodipicolinate reductase from Paenisporosarcina sp. TG-14 | | Descriptor: | Dihydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-01-02 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase (PaDHDPR) from Paenisporosarcina sp. TG-14: structural basis for NADPH preference as a cofactor
Sci Rep, 8, 2018
|
|
5Z8K
 
 | | Crystal structure of an aminotransferase in complex with product-1 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, C-6' aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hong, S.K, Cha, S.S. | | Deposit date: | 2018-01-31 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Complete reconstitution of the diverse pathways of gentamicin B biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
5Z2D
 
 | |
5XGX
 
 | | Crystal structure of colwellia psychrerythraea strain 34H isoaspartyl dipeptidase E80Q mutant complexed with beta-isoaspartyl lysine | | Descriptor: | D-ASPARTIC ACID, D-LYSINE, Isoaspartyl dipeptidase, ... | | Authors: | Lee, J.H, Lee, C.W, Park, S.H. | | Deposit date: | 2017-04-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure and functional characterization of an isoaspartyl dipeptidase (CpsIadA) from Colwellia psychrerythraea strain 34H.
PLoS ONE, 12, 2017
|
|
5Z83
 
 | |
1DC2
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 20 STRUCTURES | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR A (P16INK4A) | | Authors: | Byeon, I.-J.L, Li, J, Yuan, C, Tsai, M.-D. | | Deposit date: | 1999-11-04 | | Release date: | 1999-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: refinement of p16INK4A structure and determination of p15INK4B structure by comparative modeling and NMR data.
Protein Sci., 9, 2000
|
|
