7WGC
 
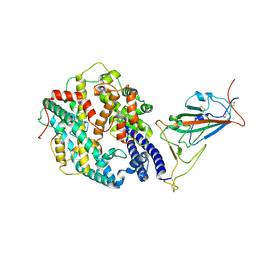 | | Neutral Omicron Spike Trimer in complex with ACE2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WG7
 
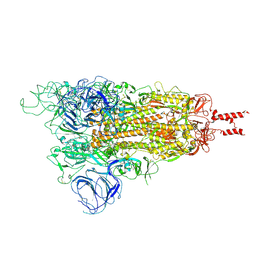 | | Acidic Omicron Spike Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WG9
 
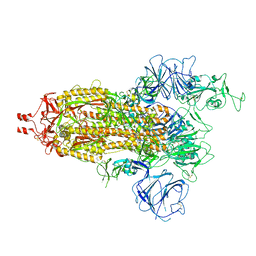 | | Delta Spike Trimer(1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WGB
 
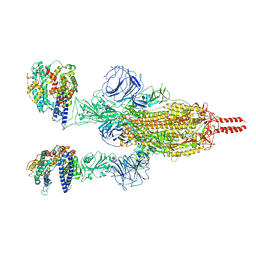 | | Neutral Omicron Spike Trimer in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WG8
 
 | | Delta Spike Trimer(3 RBD Down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7X45
 
 | | Grass carp interferon gamma related | | Descriptor: | Interferon gamma | | Authors: | Wang, J, Zou, J, Zhu, X. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Novel Dimeric Architecture of an IFN-gamma-Related Cytokine Provides Insights into Subfunctionalization of Type II IFNs in Teleost Fish.
J Immunol., 209, 2022
|
|
5Y4Z
 
 | | Crystal structure of the Zika virus NS3 helicase complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, NS3 helicase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, L. | | Deposit date: | 2017-08-06 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural view of the helicase reveals that Zika virus uses a conserved mechanism for unwinding RNA
Acta Crystallogr.,Sect.F, 74, 2018
|
|
7E3O
 
 | |
4LYP
 
 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7D9M
 
 | | grass carp interleukin-2 | | Descriptor: | Interleukin | | Authors: | Junya, w, Jun, z. | | Deposit date: | 2020-10-13 | | Release date: | 2020-10-28 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the co-evolution of IL-2 and its private receptor in fish.
Dev.Comp.Immunol., 115, 2020
|
|
7F7H
 
 | | SARS-CoV-2 S protein RBD in complex with A8-1 Fab | | Descriptor: | Heavy chain of A8-1 Fab, Light chain of A8-1 Fab, Spike glycoprotein S1 | | Authors: | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | Deposit date: | 2021-06-29 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | High throughput isolation of potent neutralizing antibodies from convalescent COVID-19 patients.
To Be Published
|
|
7F3Q
 
 | | SARS-CoV-2 RBD in complex with A5-10 Fab and A34-2 Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A34-2 Fab, ... | | Authors: | Dou, Y, Wang, X, Liu, P, Lu, B, Wang, K. | | Deposit date: | 2021-06-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Development of neutralizing antibodies against SARS-CoV-2, using a high-throughput single-B-cell cloning method.
Antib Ther, 6, 2023
|
|
7CE2
 
 | | The Crystal structure of TeNT Hc complexed with neutralizing antibody | | Descriptor: | Tetanus toxin, neutralizing antibody heavy chain, neutralizing antibody light chain | | Authors: | Wang, X, Wang, Y, Wu, C, Yu, J, Liao, H. | | Deposit date: | 2020-06-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of tetanus toxin neutralization by native human monoclonal antibodies.
Cell Rep, 35, 2021
|
|
8H7Z
 
 | |
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | Authors: | Liu, Z, Yan, A, Gao, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
3O5X
 
 | | Crystal structure of the oncogenic tyrosine phosphatase SHP2 complexed with a salicylic acid-based small molecule inhibitor | | Descriptor: | 3-{1-[3-(biphenyl-4-ylamino)-3-oxopropyl]-1H-1,2,3-triazol-4-yl}-6-hydroxy-1-methyl-2-phenyl-1H-indole-5-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.-Y, Zhang, X, He, Y, Liu, S, Yu, Z, Jiang, Z, Yang, Z, Dong, Y, Nabinger, S.C, Wu, L, Gunawan, A.M, Wang, L, Chan, R.J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2).
J.Med.Chem., 53, 2010
|
|
7BUJ
 
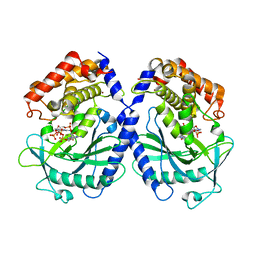 | | mcGAS bound with pppGpG | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, B, Su, X.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Mn2+Directly Activates cGAS and Structural Analysis Suggests Mn2+Induces a Noncanonical Catalytic Synthesis of 2'3'-cGAMP.
Cell Rep, 32, 2020
|
|
7BUM
 
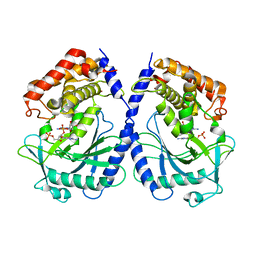 | | mcGAS bound with pGpA | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Wang, B, Su, X.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Mn2+Directly Activates cGAS and Structural Analysis Suggests Mn2+Induces a Noncanonical Catalytic Synthesis of 2'3'-cGAMP.
Cell Rep, 32, 2020
|
|
7BUQ
 
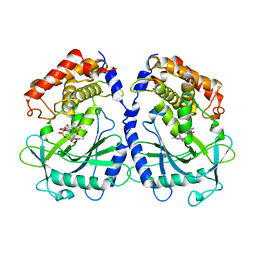 | | mcGAS bound with 23-cGAMP | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION, cGAMP | | Authors: | Wang, B, Su, X.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Mn2+Directly Activates cGAS and Structural Analysis Suggests Mn2+Induces a Noncanonical Catalytic Synthesis of 2'3'-cGAMP.
Cell Rep, 32, 2020
|
|
7RMR
 
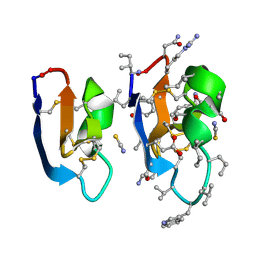 | | Crystal structure of [I11L]cycloviolacin O2 | | Descriptor: | D-[I11L]cycloviolacin O2, THIOCYANATE ION, [I11L]cycloviolacin O2 | | Authors: | Huang, Y.H, Du, Q, Craik, D.J. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
7RMQ
 
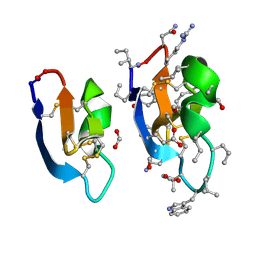 | | Crystal structure of cycloviolacin O2 | | Descriptor: | Cycloviolacin O2, D-[I11L]cycloviolacin O2, FORMIC ACID | | Authors: | Huang, Y.H, Du, Q. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
7RMS
 
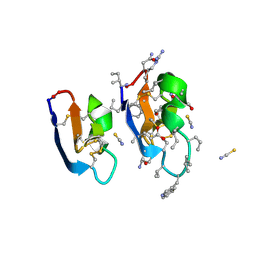 | | Crystal structure of [I11G]cycloviolacin O2 | | Descriptor: | D-[I11L]cycloviolacin O2, THIOCYANATE ION, [I11L]cycloviolacin O2 | | Authors: | Huang, Y.H, Du, Q, Craik, D.J. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
4LYR
 
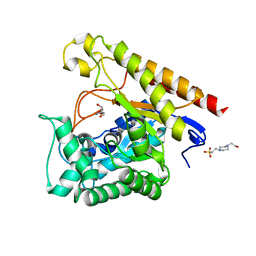 | | Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E301A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LYQ
 
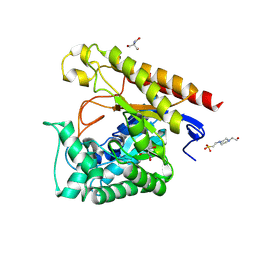 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Exo-beta-1,4-mannosidase, ... | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NRS
 
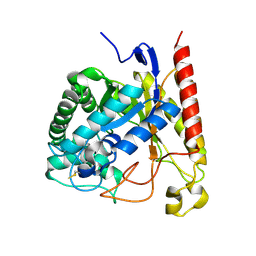 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose | | Descriptor: | Exo-beta-1,4-mannosidase, beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-11-27 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
