3LQK
 
 | | Crystal structure of dipicolinate synthase subunit B from Bacillus halodurans C | | Descriptor: | Dipicolinate synthase subunit B, PHOSPHATE ION | | Authors: | Nocek, B, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-09 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dipicolinate synthase subunit B from Bacillus halodurans C
To be Published
|
|
1LXN
 
 | | X-RAY STRUCTURE OF MTH1187 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT272 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1187, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of MTH1187 and its Yeast Ortholog YBL001C
Proteins, 52, 2003
|
|
1T9K
 
 | | X-ray crystal structure of aIF-2B alpha subunit-related translation initiation factor [Thermotoga maritima] | | Descriptor: | CHLORIDE ION, Probable methylthioribose-1-phosphate isomerase, SULFATE ION | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-17 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystal structure of aIF-2B translation initiation factor from Thermotoga maritima
To be Published
|
|
1S5U
 
 | | Crystal Structure of Hypothetical Protein EC709 from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Protein ybgC, SULFATE ION | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-21 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Hypothetical Protein EC709 from Escherichia coli
To be Published
|
|
1S7I
 
 | | 1.8 A Crystal Structure of a Protein of Unknown Function PA1349 from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA1349 | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-29 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8A crystal structure of a hypothetical protein PA1349 from Pseudomonas aeruginosa
To be Published
|
|
5VRA
 
 | | 2.35-Angstrom In situ Mylar structure of human A2A adenosine receptor at 100 K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Broecker, J, Morizumi, T, Ou, W.-L, Klingel, V, Kuo, A, Kissick, D.J, Ishchenko, A, Lee, M.-Y, Xu, S, Makarov, O, Cherezov, V, Ogata, C.M, Ernst, O.P. | | Deposit date: | 2017-05-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
1SED
 
 | | Crystal Structure of Protein of Unknown Function YhaL from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Hypothetical protein yhaI, ... | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-17 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Hypothetical Protein YhaI, APC1180 from Bacillus subtilis
To be Published
|
|
1LXJ
 
 | | X-RAY STRUCTURE OF YBL001c NORTHEAST STRUCTURAL GENOMICS (NESG) CONSORTIUM TARGET YTYst72 | | Descriptor: | HYPOTHETICAL 11.5KDA PROTEIN IN HTB2-NTH2 INTERGENIC REGION, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURES OF MTH1187 AND ITS YEAST ORTHOLOG YBL001C
Proteins, 52, 2003
|
|
1SQU
 
 | |
1K77
 
 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | Descriptor: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | Authors: | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-18 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
3LVY
 
 | | Crystal Structure of Carboxymuconolactone Decarboxylase Family Protein SMU.961 from Streptococcus mutans | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Carboxymuconolactone decarboxylase family, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Carboxymuconolactone Decarboxylase Family Protein SMU.961 from Streptococcus mutans
To be Published
|
|
8EFZ
 
 | | Crystal structure of CcNikZ-II, apoprotein | | Descriptor: | CHLORIDE ION, Extracellular solute-binding protein family 5 | | Authors: | Stogios, P.J, Evdokimova, E, Diep, P, Yakunin, A, Mahadevan, K, Savchenko, A. | | Deposit date: | 2022-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Ni(II)-binding affinity of CcNikZ-II and its homologs: the role of the HH-prong and variable loop revealed by structural and mutational studies.
Febs J., 291, 2024
|
|
3LZK
 
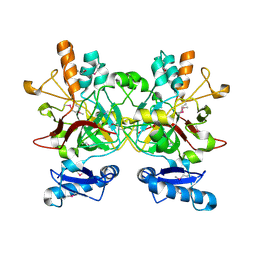 | | The crystal structure of a probably aromatic amino acid degradation proteiN from Sinorhizobium meliloti 1021 | | Descriptor: | CALCIUM ION, Fumarylacetoacetate hydrolase family protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a probably aromatic amino acid degradation protein from Sinorhizobium meliloti 1021
To be Published
|
|
4WZ0
 
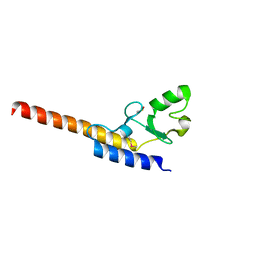 | | Crystal structure of U-box 1 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris | | Descriptor: | E3 ubiquitin-protein ligase LubX | | Authors: | Stogios, P.J, Quaile, A.T, Skarina, T, Stein, A, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
6XYN
 
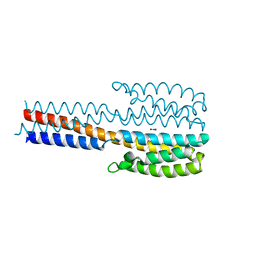 | | Crystal structure of a proteolytic fragment of NarQ comprising sensor and TM domains | | Descriptor: | NITRATE ION, Nitrate/nitrite sensor histidine kinase NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Gordeliy, V. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a proteolytic fragment of the sensor histidine kinase NarQ
Crystals, 2020
|
|
3MT1
 
 | | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti | | Descriptor: | Putative carboxynorspermidine decarboxylase protein, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti
To be Published
|
|
1SR8
 
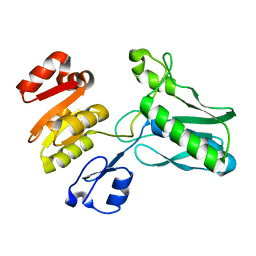 | | Structural Genomics, 1.9A crystal structure of cobalamin biosynthesis protein (cbiD) from Archaeoglobus fulgidus | | Descriptor: | cobalamin biosynthesis protein (cbiD) | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-22 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9A crystal structure of cobalamin biosynthesis protein (cbiD) from Archaeoglobus fulgidus
To be Published
|
|
3MQZ
 
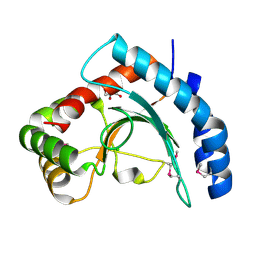 | | Crystal Structure of Conserved Protein DUF1054 from Pink Subaerial Biofilm Microbial Leptospirillum sp. Group II UBA. | | Descriptor: | CHLORIDE ION, GLYCEROL, uncharacterized Conserved Protein DUF1054 | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Conserved Protein DUF1054 from Pink Subaerial Biofilm Microbial Leptospirillum sp. Group II UBA.
To be Published
|
|
3M6J
 
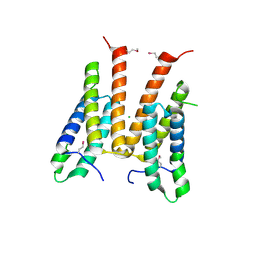 | | Crystal structure of unknown function protein from Leptospirillum rubarum | | Descriptor: | CHLORIDE ION, uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-15 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of unknown function protein from Leptospirillum rubarum
To be Published
|
|
5BY0
 
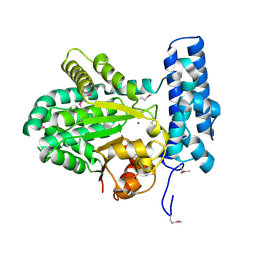 | | Crystal structure of magnesium-bound Duf89 protein Saccharomyces cerevisiae | | Descriptor: | MAGNESIUM ION, Protein-glutamate O-methyltransferase | | Authors: | Nocek, B, Cuff, M, Cui, H, Xu, X, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of magnesium-bound Duf89 protein Saccharomyces cerevisiae
To Be Published
|
|
3M92
 
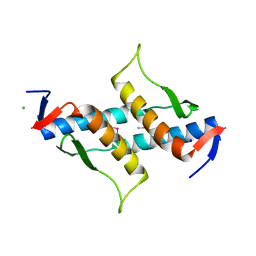 | | The structure of yciN, an unchracterized protein from Shigella flexneri. | | Descriptor: | CHLORIDE ION, Protein yciN, SODIUM ION | | Authors: | Cuff, M.E, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-19 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of yciN, an unchracterized protein from Shigella flexneri.
TO BE PUBLISHED
|
|
4Q3O
 
 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
3MOI
 
 | | The crystal structure of the putative dehydrogenase from Bordetella bronchiseptica RB50 | | Descriptor: | Probable dehydrogenase | | Authors: | Zhang, R, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the putative dehydrogenase from Bordetella bronchiseptica RB50
To be Published
|
|
1U60
 
 | | MCSG APC5046 Probable glutaminase ybaS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable glutaminase ybaS | | Authors: | Chang, C, Cuff, M.E, Joachimiak, A, Savchenko, A, Edwards, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
5ES2
 
 | | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To Be Published
|
|
