7U6K
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7UJI
 
 | |
7UOG
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and asymmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence. | | Descriptor: | DNA (5'-D(AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*CP*CP*GP*CP*CP*TP*T)-3', DNA (5'-D(GP*TP*AP*AP*GP*GP*CP*GP*GP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*G)-3', MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7UR0
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and scaffold-insert duplexes (21mer and 10mer) containing the cognate REPE54 sequence and an additional T-A rich sequence, respectively. | | Descriptor: | DNA (5'-D(*AP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*A)-3'), DNA (5'-D(A*GP*AP*CP*GP*GP*TP*AP*AP*TP*T)-3'), DNA (5'-D(A*GP*TP*AP*AP*TP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-04-20 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
8SP7
 
 | |
7ZBB
 
 | | HaloTag with TRaQ-G-ctrl ligand | | Descriptor: | (E)-[7-azanyl-10-[2-carboxy-5-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]phenyl]-5,5-dimethyl-benzo[b][1]benzosilin-3-ylidene]-methyl-azanium, CHLORIDE ION, GLYCEROL, ... | | Authors: | Emmert, S, Rivera-Fuentes, P, Pojer, F, Lau, K. | | Deposit date: | 2022-03-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A locally activatable sensor for robust quantification of organellar glutathione.
Nat.Chem., 15, 2023
|
|
7ZBA
 
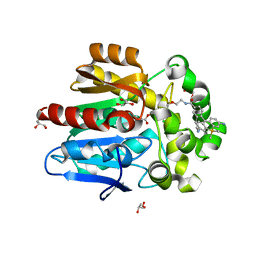 | | HaloTag with Me-TRaQ-G ligand | | Descriptor: | 4-(7-azanyl-5,5-dimethyl-3-methylimino-benzo[b][1]benzosilin-10-yl)-N-[2-[2-(6-chloranylhexoxy)ethoxy]ethyl]-3-methyl-benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Emmert, S, Rivera-Fuentes, P, Pojer, F, Lau, K. | | Deposit date: | 2022-03-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | A locally activatable sensor for robust quantification of organellar glutathione.
Nat.Chem., 15, 2023
|
|
7ZBD
 
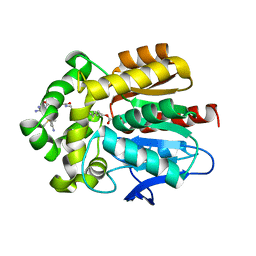 | | HaloTag with TRaQ-G ligand | | Descriptor: | (10R)-7-azanyl-N-[2-[2-(6-chloranylhexoxy)ethoxy]ethyl]-2'-cyano-5,5-dimethyl-3-(methylamino)-1'-oxidanylidene-spiro[benzo[b][1]benzosiline-10,3'-isoindole]-5'-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Emmert, S, Rivera-Fuentes, P, Pojer, F, Lau, K. | | Deposit date: | 2022-03-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A locally activatable sensor for robust quantification of organellar glutathione.
Nat.Chem., 15, 2023
|
|
7UXY
 
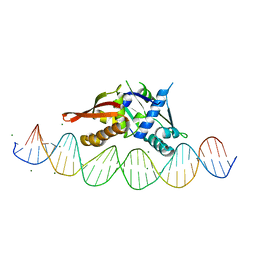 | | Isoreticular, interpenetrating co-crystal of protein variant Replication Initiator Protein REPE54 (L53G,Q54G,E55G) and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*P*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*P*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
8AHW
 
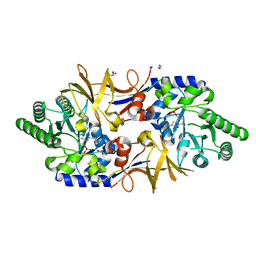 | | Structure of DCS-resistant variant D322N of alanine racemase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Alanine racemase, GLYCEROL | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-07-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
8PXI
 
 | | Crystal structure of Endothiapepsin soaked with FRG283 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-{[methyl(prop-2-yn-1-yl)amino]methyl}-1,3-thiazol-4-yl)piperidin-4-ol, DIMETHYL SULFOXIDE, ... | | Authors: | Mueller, J.M, Eckelt, S, Klebe, G, Glinca, S. | | Deposit date: | 2023-07-23 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of Endothiapepsin with ligands derived from merged fragment hits
To Be Published
|
|
9ONO
 
 | | Fe-bound B. pseudomallei rubrerythrin | | Descriptor: | FE (III) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9NND
 
 | | Structure of the HERV-K (HML-2) spike complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaked, R, Katz, M, Diskin, R. | | Deposit date: | 2025-03-05 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | The prefusion structure of the HERV-K spike complex
To Be Published
|
|
9NYO
 
 | |
9R2L
 
 | | De novo designed N37 protein fold | | Descriptor: | ACETATE ION, ETHANOLAMINE, N37, ... | | Authors: | Pacesa, M, Miao, Y, Georgeon, S, Schmidt, J, Correia, B.E. | | Deposit date: | 2025-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reimagine accessibility of protein space with generative model
To Be Published
|
|
9R2R
 
 | |
9R2V
 
 | | De novo designed M16 protein fold | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, FORMIC ACID, ... | | Authors: | Pacesa, M, Miao, Y, Georgeon, S, Schmidt, J, Correia, B.E. | | Deposit date: | 2025-05-01 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Reimagine accessibility of protein space with generative model
To Be Published
|
|
9ONR
 
 | | rubrerythrin- E53A E124A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Rubrerythrin, THIOCYANATE ION | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONN
 
 | | Co-bound B. pseudomallei Rubrerythrin | | Descriptor: | COBALT (II) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONM
 
 | | near-apo rubrerythrin | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONQ
 
 | | Mn-bound B. pseudomallei rubrerythrin | | Descriptor: | MANGANESE (II) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9R2O
 
 | | De novo designed N5 protein fold | | Descriptor: | N5, POTASSIUM ION, THIOCYANATE ION | | Authors: | Pacesa, M, Miao, Y, Georgeon, S, Schmidt, J, Correia, B.E. | | Deposit date: | 2025-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Reimagine accessibility of protein space with generative model
To Be Published
|
|
9QMP
 
 | | E.coli seryl-tRNA synthetase | | Descriptor: | Serine--tRNA ligase | | Authors: | Cusack, S. | | Deposit date: | 2025-03-24 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A.
Nature, 347, 1990
|
|
7SGC
 
 | |
7SDP
 
 | |
