6NTS
 
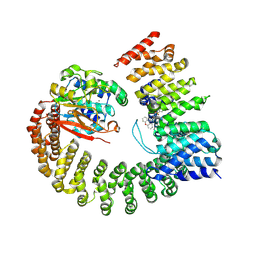 | | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | | Descriptor: | MANGANESE (II) ION, N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2019-01-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Cell, 181, 2020
|
|
8H5Z
 
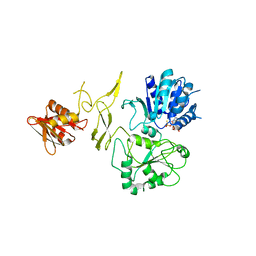 | | Crystal structure of RadD/ATP analogue complex | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Putative DNA repair helicase RadD, ZINC ION | | Authors: | Yan, X.X, Tian, L.F. | | Deposit date: | 2022-10-14 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.00002718 Å) | | Cite: | Biochemical and Structural Analyses Shed Light on the Mechanisms of RadD DNA Binding and Its ATPase from Escherichia coli.
Int J Mol Sci, 24, 2023
|
|
8H5Y
 
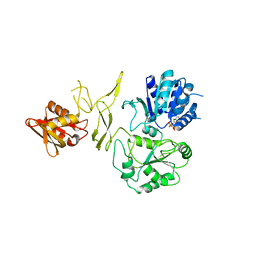 | | Crystal structure of RadD- ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative DNA repair helicase RadD, ... | | Authors: | Yan, X.X, Tian, L.F. | | Deposit date: | 2022-10-14 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7001 Å) | | Cite: | Biochemical and Structural Analyses Shed Light on the Mechanisms of RadD DNA Binding and Its ATPase from Escherichia coli.
Int J Mol Sci, 24, 2023
|
|
4XAU
 
 | | Crystal structure of AtS13 from Actinomadura melliaura | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative aminotransferase | | Authors: | Wang, F, Singh, S, Xu, W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0012 Å) | | Cite: | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
5CMZ
 
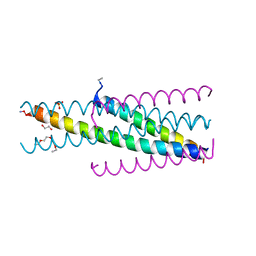 | | Artificial HIV fusion inhibitor AP3 fused to the C-terminus of gp41 NHR | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Artificial HIV entry inhibitor AP3, ... | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
6D2L
 
 | | Crystal structure of human CARM1 with (S)-SKI-72 | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-hydroxyphenyl)ethyl]hexanamide, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Hutchinson, A, Seitova, A, LUO, M, CAI, X.C, KE, W, WANG, J, SHI, C, ZHENG, W, LEE, J.P, IBANEZ, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
5CN0
 
 | | Artificial HIV fusion inhibitor AP2 fused to the C-terminus of gp41 NHR | | Descriptor: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein,AP2, MAGNESIUM ION | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CMU
 
 | |
2KPH
 
 | | NMR Structure of AtraPBP1 at pH 4.5 | | Descriptor: | Pheromone binding protein | | Authors: | Ames, J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Navel Orangeworm Moth Pheromone-Binding Protein (AtraPBP1): Implications for pH-Sensitive Pheromone Detection .
Biochemistry, 49, 2010
|
|
7PRQ
 
 | | Structure of the ligand binding domain of the PctD (PA4633) chemoreceptor of Pseudomonas aeruginosa PAO1 in complex with choline. | | Descriptor: | 1,2-ETHANEDIOL, CHOLINE ION, GLYCEROL, ... | | Authors: | Gavira, J.A, Matilla, M.A, Martin-Mora, D, Krell, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
7PSG
 
 | | Structure of the ligand binding domain of the PacA (ECA2226) chemoreceptor of Pectobacterium atrosepticum SCRI1043 in complex with betaine. | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, TRIMETHYL GLYCINE | | Authors: | Gavira, J.A, Matilla, M.A, Velando, F, Krell, T. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
7PRR
 
 | | Structure of the ligand binding domain of the PctD (PA4633) chemoreceptor of Pseudomonas aeruginosa PAO1 in complex with acetylcholine | | Descriptor: | ACETYLCHOLINE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Matilla, M.A, Martin-Mora, D, Krell, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemotaxis of the Human Pathogen Pseudomonas aeruginosa to the Neurotransmitter Acetylcholine.
Mbio, 13, 2022
|
|
3OGN
 
 | |
1QA0
 
 | | BOVINE TRYPSIN 2-AMINOBENZIMIDAZOLE COMPLEX | | Descriptor: | 2H-BENZOIMIDAZOL-2-YLAMINE, CALCIUM ION, TRYPSIN | | Authors: | Whitlow, M. | | Deposit date: | 1999-04-09 | | Release date: | 2000-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of potent and selective factor Xa inhibitors complexed to bovine trypsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4IKG
 
 | |
4IKP
 
 | | Crystal structure of coactivator-associated arginine methyltransferase 1 with methylenesinefungin | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Dombrovski, L, He, H, Ibanez, G, Wernimont, A, Zheng, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-27 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
4OLT
 
 | | Chitosanase complex structure | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Chitosanase, GLYCEROL | | Authors: | Liu, W.Z, Lyu, Q.Q, Han, B.Q. | | Deposit date: | 2014-01-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into the substrate-binding mechanism for a novel chitosanase.
Biochem.J., 461, 2014
|
|
5UWG
 
 | | The crystal structure of Fz4-CRD in complex with palmitoleic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, PALMITOLEIC ACID | | Authors: | Ke, J, DeBruine, Z.J, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Wnt5a promotes Frizzled-4 signalosome assembly by stabilizing cysteine-rich domain dimerization.
Genes Dev., 31, 2017
|
|
3B6Z
 
 | | Lovastatin polyketide enoyl reductase (LovC) complexed with 2'-phosphoadenosyl isomer of crotonoyl-CoA | | Descriptor: | Enoyl reductase, GLYCEROL, S-{(9R,13R,15S)-17-[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]-9,13,15-trihydroxy-10,10-dimethyl-13,15-dioxido-4,8-dioxo-12,14,16-trioxa-3,7-diaza-13,15-diphosphaheptadec-1-yl}(2E)-but-2-enethioate | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B70
 
 | | Crystal structure of Aspergillus terreus trans-acting lovastatin polyketide enoyl reductase (LovC) with bound NADP | | Descriptor: | Enoyl reductase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6B85
 
 | | Crystal structure of transmembrane protein TMHC4_R | | Descriptor: | TMHC4_R | | Authors: | Lu, P, DiMaio, F, Min, D, Bowie, J, Wei, K.Y, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.889 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6B87
 
 | | Crystal structure of transmembrane protein TMHC2_E | | Descriptor: | TMHC2_E | | Authors: | Lu, P, DiMaio, F, Min, D, Wei, K.Y, Bowie, J, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6CCM
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 2-((3-bromobenzyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one | | Descriptor: | 2-{[(3-bromophenyl)methyl]amino}-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6CCS
 
 | |
6CHN
 
 | | Phosphopantetheine adenylyltransferase (CoaD) in complex with methyl (R)-4-(3-(2-cyano-1-((5-methyl-7-oxo-4,7-dihydro-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino)ethyl)phenoxy)piperidine-1-carboxylate | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosphopantetheine adenylyltransferase, SULFATE ION, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and Optimization of Phosphopantetheine Adenylyltransferase Inhibitors with Gram-Negative Antibacterial Activity.
J. Med. Chem., 61, 2018
|
|
