8KG8
 
 | | Yeast replisome in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8K14
 
 | | X-ray crystal structure of 18a in BRD4(1) | | Descriptor: | 4-[8-methoxy-2-methyl-1-(1-phenylethyl)imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Xu, H, Shen, H, Zhang, Y, Xu, Y, Li, R. | | Deposit date: | 2023-07-10 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of (R)-4-(8-methoxy-2-methyl-1-(1-phenylethy)-1H-imidazo[4,5-c]quinnolin-7-yl)-3,5-dimethylisoxazole as a potent and selective BET inhibitor for treatment of acute myeloid leukemia (AML) guided by FEP calculation.
Eur.J.Med.Chem., 263, 2024
|
|
8KG9
 
 | | Yeast replisome in state III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
4GR7
 
 | | The human W42R Gamma D-Crystallin Mutant Structure at 1.7A Resolution | | Descriptor: | Gamma-crystallin D, PHOSPHATE ION | | Authors: | Ji, F, Jung, J, Koharudin, L.M.I, Gronenborn, A.M. | | Deposit date: | 2012-08-24 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The human W42R gamma D-crystallin mutant structure provides a link between congenital and age-related cataracts.
J.Biol.Chem., 288, 2013
|
|
7WVM
 
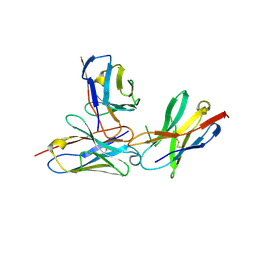 | | The complex structure of PD-1 and cemiplimab | | Descriptor: | Heavy Chain of Cemiplimab, Light Chain of Cemiplimab, Programmed cell death protein 1 | | Authors: | Lu, D, Xu, Z.P, Liu, K.F, Tan, S.G, Gao, G.F, Chai, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | PD-1 N58-Glycosylation-Dependent Binding of Monoclonal Antibody Cemiplimab for Immune Checkpoint Therapy.
Front Immunol, 13, 2022
|
|
7WLI
 
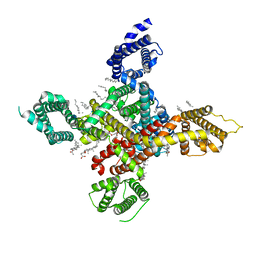 | | CryoEM structure of human low-voltage activated T-type calcium channel CaV3.3 (apo) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLK
 
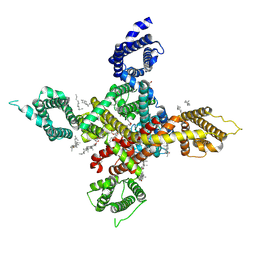 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with Otilonium Bromide(OB) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-[diethyl(methyl)-$l^{4}-azanyl]ethyl 4-[(2-octoxyphenyl)carbonylamino]benzoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLJ
 
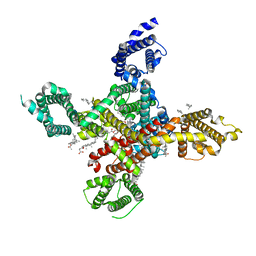 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with mibefradil (MIB) | | Descriptor: | (1S,2S)-2-(2-{[3-(1H-benzimidazol-2-yl)propyl](methyl)amino}ethyl)-6-fluoro-1-(propan-2-yl)-1,2,3,4-tetrahydronaphthalen-2-yl methoxyacetate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLL
 
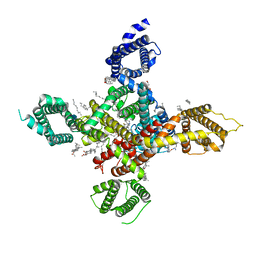 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with pimozide(PMZ) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7T5I
 
 | | Crystal Structure of Strictosidine Synthase in complex with R-IPA(2-(1H-indol-3-yl) propan-1-amine) | | Descriptor: | (2R)-2-(1H-indol-3-yl)propan-1-amine, Strictosidine synthase | | Authors: | Liu, H, Panjikar, S, Futamura, Y, Shao, N, Osada, H, Zou, H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Beta-Branched Tryptamine Provoked Non Sandwich-Like-Mode Catalysis of Strictosidine Synthase to Antimalarial Indole Alkaloids
to be published
|
|
7T5J
 
 | | Crystal Structure of Strictosidine Synthase in complex with S-IPA (2-(1H-indol-3-yl) propan-1-amine) | | Descriptor: | (2S)-2-(1H-indol-3-yl)propan-1-amine, Strictosidine synthase | | Authors: | Liu, H, Panjikar, S, Futamura, Y, Shao, N, Osada, H, Zou, H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | beta-Branched Tryptamine Provoked Non Sandwich-Like-Mode Catalysis of Strictosidine Synthase to Antimalarial Indole Alkaloids
to be published
|
|
7CR6
 
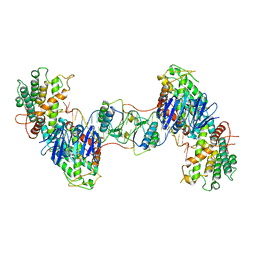 | | Synechocystis Cas1-Cas2/prespacer binary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
7CSD
 
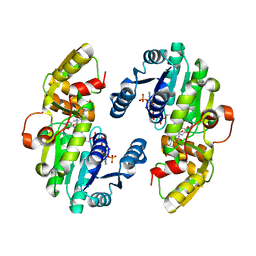 | | AtPrR1 with NADP+ and (+)lariciresinol | | Descriptor: | 4-[[(3R,4R,5S)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79709971 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS2
 
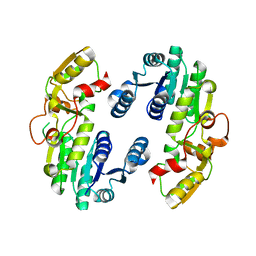 | | Apo structure of dimeric IiPLR1 | | Descriptor: | Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68800473 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSF
 
 | | AtPrR1 with NADP+ and (-)secoisolariciresinol | | Descriptor: | (2R,3R)-2,3-bis[(3-methoxy-4-oxidanyl-phenyl)methyl]butane-1,4-diol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98241806 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS3
 
 | | IiPLR1 with NADP+ | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.40021849 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS6
 
 | | IiPLR1 with NADP+ and (-)lariciresinol | | Descriptor: | 4-[[(3S,4S,5R)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20139647 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS8
 
 | | IiPLR1 with NADP+ and (-)secoisolariciresinol | | Descriptor: | (2R,3R)-2,3-bis[(3-methoxy-4-oxidanyl-phenyl)methyl]butane-1,4-diol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.60003257 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSE
 
 | | AtPrR1 with NADP+ and (-)lariciresinol | | Descriptor: | 4-[[(3S,4S,5R)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44019055 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS4
 
 | | IiPLR1 with NADP+ and (+)pinoresinol | | Descriptor: | 4-[(3S,3aR,6S,6aR)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.30509257 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSC
 
 | | AtPrR1 with NADP+ and (-)pinoresinol | | Descriptor: | 4-[(3R,3aS,6R,6aS)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5151186 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSB
 
 | | AtPrR1 with NADP+ and (+)pinoresinol | | Descriptor: | 4-[(3S,3aR,6S,6aR)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.002017 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS5
 
 | | IiPLR1 with NADP+ and (-)pinoresinol | | Descriptor: | 4-[(3R,3aS,6R,6aS)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18993235 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSH
 
 | | AtPrR2 with NADP+ and (+)pinoresinol | | Descriptor: | 4-[(3S,3aR,6S,6aR)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 2 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.590775 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS9
 
 | | AtPrR1 in apo form | | Descriptor: | Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8011415 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
