2EFI
 
 | | Solution structure of the chromo domain of Mortality factor 4-like protein 1 from human | | Descriptor: | Mortality factor 4-like protein 1 | | Authors: | Li, H, Sato, M, Tochio, N, Tomizawa, T, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chromo domain of Mortality factor 4-like protein 1 from human
To be Published
|
|
2FSA
 
 | |
2F6J
 
 | |
2F6N
 
 | |
2D9U
 
 | | Solution structure of the Chromo domain of chromobox homolog 2 from human | | Descriptor: | Chromobox protein homolog 2 (isoform 2) | | Authors: | Li, H, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Chromo domain of chromobox homolog 2 from human
To be Published
|
|
2D9Z
 
 | | Solution structure of the PH domain of Protein kinase C, nu type from human | | Descriptor: | Protein kinase C, nu type | | Authors: | Li, H, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of Protein kinase C, nu type from human
To be Published
|
|
2D9V
 
 | | Solution structure of the PH domain of Pleckstrin homology domain-containing protein family B member 1 from mouse | | Descriptor: | Pleckstrin homology domain-containing protein family B member 1 | | Authors: | Li, H, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of Pleckstrin homology domain-containing protein family B member 1 from mouse
To be Published
|
|
2D9X
 
 | | Solution structure of the PH domain of Oxysterol binding protein-related protein 11 from human | | Descriptor: | Oxysterol binding protein-related protein 11 | | Authors: | Li, H, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of Oxysterol binding protein-related protein 11 from human
To be Published
|
|
2E6J
 
 | | Solution structure of the C-terminal PapD-like domain from human HYDIN protein | | Descriptor: | HYDIN protein | | Authors: | Li, H, Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-27 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal PapD-like domain from human HYDIN protein
To be Published
|
|
6JAT
 
 | | Crystal structure of SETD3 bound to Actin peptide and SFG | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Actin, gamma-enteric smooth muscle, ... | | Authors: | Li, H, Zheng, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | Crystal structure of SETD3 bound to Actin peptide and SFG
to be published
|
|
8H7P
 
 | | Crystal structure of aqualigase bound with Suc-AAPF | | Descriptor: | 5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, CALCIUM ION, Subtilisin, ... | | Authors: | Li, H, Ma, M.Z, Zhang, L.J, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of aqualigase bound with Suc-AAPF
To Be Published
|
|
8H7O
 
 | | Crystal structure of aqualigase | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, Subtilisin | | Authors: | Li, H, Ma, M.Z, Zhang, L.J, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of aqualigase
To Be Published
|
|
8J8I
 
 | |
8IGL
 
 | |
8JZ6
 
 | |
5ZGC
 
 | | Crystal Structure of SIRT3 in complex with H4K16bhb peptide | | Descriptor: | Histone H4K16bhb peptide, NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ... | | Authors: | Li, H, Zhang, X. | | Deposit date: | 2018-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of SIRT3 in complex with H4K16bhb peptide
To be published
|
|
6JIT
 
 | | Complex structure of an imine reductase at 2.05 Angstrom resolution | | Descriptor: | 1-(2-phenylethyl)-3,4-dihydroisoquinoline, 6-phosphogluconate dehydrogenase NAD-binding protein, CHLORIDE ION, ... | | Authors: | Li, H, Wu, L, Zheng, G.W, Zhou, J.H. | | Deposit date: | 2019-02-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Complex structure of an imine reductase at 2.05 Angstrom resolution
To Be Published
|
|
6JIZ
 
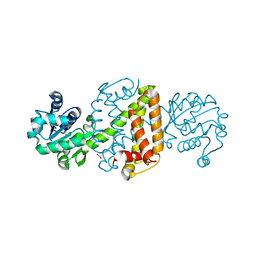 | | Apo structure of an imine reductase at 1.76 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 3-ethylheptane, 6-phosphogluconate dehydrogenase NAD-binding protein, ... | | Authors: | Li, H, Wu, L, Zheng, G.W, Zhou, J.H. | | Deposit date: | 2019-02-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Apo structure of an imine reductase at 1.76 Angstrom resolution
To Be Published
|
|
7W1W
 
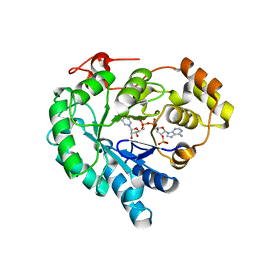 | | NADPH-bound AKR4C17 mutant F291D | | Descriptor: | AKR4-2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-11-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7W1X
 
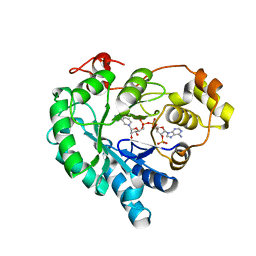 | | Crystal structure of AKR4C16 bound with NADPH | | Descriptor: | AKR4-1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-11-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7UOT
 
 | | Native Lassa glycoprotein in complex with neutralizing antibodies 8.9F and 37.2D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 37.2D heavy chain (variable domain), ... | | Authors: | Li, H, Saphire, E.O. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | A cocktail of protective antibodies subverts the dense glycan shield of Lassa virus.
Sci Transl Med, 14, 2022
|
|
7UOV
 
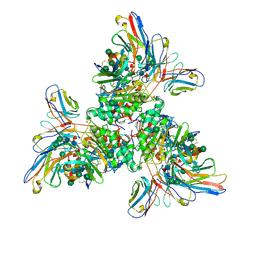 | |
9AWJ
 
 | | Bovine adult muscle nAChR bound to ACh | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE, ... | | Authors: | Li, H, Hibbs, R.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural switch in acetylcholine receptors in developing muscle.
Nature, 632, 2024
|
|
9AVU
 
 | | Bovine fetal muscle nAChR bound to ACh | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE, ... | | Authors: | Li, H, Hibbs, R.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural switch in acetylcholine receptors in developing muscle.
Nature, 632, 2024
|
|
9AVV
 
 | | Bovine adult muscle nAChR resting state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine receptor subunit alpha, ... | | Authors: | Li, H, Hibbs, R.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Structural switch in acetylcholine receptors in developing muscle.
Nature, 632, 2024
|
|
