7VDH
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80, state2 | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VDM
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with substance P | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
6IUF
 
 | | Crystal structure of Anti-CRISPR protein AcrVA5 | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, protein-a | | Authors: | Dong, L, Guan, X, Zhu, Y, Huang, Z. | | Deposit date: | 2018-11-28 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | An anti-CRISPR protein disables type V Cas12a by acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
7VUZ
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with PAMP-12, state2 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VV5
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80, state1 | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VV4
 
 | |
7VV0
 
 | |
7VV6
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80 (local) | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Mas-related G-protein coupled receptor member X2 | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VUY
 
 | |
7VV3
 
 | |
6KXS
 
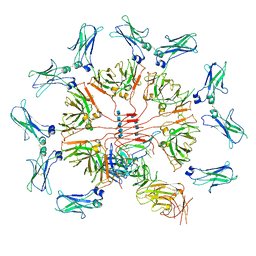 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the ectodomain of pIgR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Li, Y, Wang, G, Xiao, J. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into immunoglobulin M.
Science, 367, 2020
|
|
4J3D
 
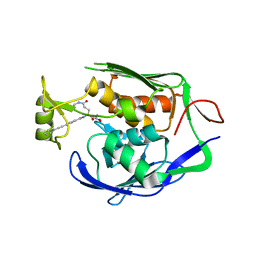 | | Pseudomonas aeruginosa LpxC in complex with a hydroxamate inhibitor | | Descriptor: | N~1~-hydroxy-N~5~-(3-hydroxypropyl)-N~2~-[4-(phenylethynyl)benzoyl]-L-glutamamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Lahiri, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the UDP pocket of LpxC through amino acid analogs.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8ZP4
 
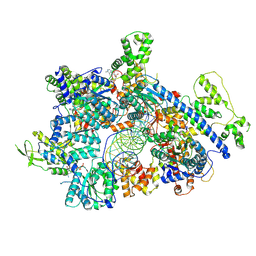 | | Cryo-EM structure of origin recognition complex (Orc1 to 5) with ARS1 DNA bound | | Descriptor: | DNA (31-MER), MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Lam, W.H, Yu, D, Dang, S, Zhai, Y. | | Deposit date: | 2024-05-29 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | DNA bending mediated by ORC is essential for replication licensing in budding yeast.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8ZPK
 
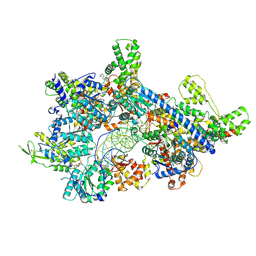 | | Cryo-EM structure of origin recognition complex (Orc6 with residues 1 to 270 deleted) with ARS1 DNA bound | | Descriptor: | DNA (38-MER), DNA (40-MER), MAGNESIUM ION, ... | | Authors: | Lam, W.H, Yu, D, Dang, S, Zhai, Y. | | Deposit date: | 2024-05-30 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | DNA bending mediated by ORC is essential for replication licensing in budding yeast.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8ZP5
 
 | | Cryo-EM structure of origin recognition complex (Orc5 basic patch mutations) with ARS1 DNA bound | | Descriptor: | DNA (34-MER), DNA (35-MER), MAGNESIUM ION, ... | | Authors: | Lam, W.H, Yu, D, Dang, S, Zhai, Y. | | Deposit date: | 2024-05-29 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | DNA bending mediated by ORC is essential for replication licensing in budding yeast.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
5Y4O
 
 | | Cryo-EM structure of MscS channel, YnaI | | Descriptor: | Low conductance mechanosensitive channel YnaI | | Authors: | Zhang, Y, Yu, J. | | Deposit date: | 2017-08-04 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A binding-block ion selective mechanism revealed by a Na/K selective channel.
Protein Cell, 9, 2018
|
|
7YLA
 
 | | Cryo-EM structure of 50S-HflX complex | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Damu, W, Ning, G. | | Deposit date: | 2022-07-25 | | Release date: | 2023-01-04 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM Structure of the 50S-HflX Complex Reveals a Novel Mechanism of Antibiotic Resistance in E. coli
Biorxiv, 2022
|
|
8HBN
 
 | | Structure of the Mex67-Mtr2-1 heterodimer | | Descriptor: | mRNA export factor MEX67, mRNA transport regulator MTR2 | | Authors: | Li, Z.Q, Chen, S.J, Sui, S.F. | | Deposit date: | 2022-10-29 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Nuclear export of pre-60S particles through the nuclear pore complex.
Nature, 618, 2023
|
|
8HFR
 
 | | NPC-trapped pre-60S particle | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Z.Q, Chen, S.J, Sui, S.F. | | Deposit date: | 2022-11-12 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Nuclear export of pre-60S particles through the nuclear pore complex.
Nature, 618, 2023
|
|
7BP3
 
 | | Cryo-EM structure of the human MCT2 | | Descriptor: | Monocarboxylate transporter 2 | | Authors: | Zhang, B, Jin, Q, Zhang, X, Guo, J, Ye, S. | | Deposit date: | 2020-03-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cooperative transport mechanism of human monocarboxylate transporter 2.
Nat Commun, 11, 2020
|
|
6J5W
 
 | | Ligand-triggered allosteric ADP release primes a plant NLR complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Disease resistance RPP13-like protein 4, RKS1 | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand-triggered allosteric ADP release primes a plant NLR complex.
Science, 364, 2019
|
|
6J5U
 
 | | Ligand-triggered allosteric ADP release primes a plant NLR complex | | Descriptor: | Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, Protein kinase superfamily protein, ... | | Authors: | Wang, J.Z, Wang, J, Meijuan, H, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ligand-triggered allosteric ADP release primes a plant NLR complex.
Science, 364, 2019
|
|
6J5V
 
 | | Ligand-triggered allosteric ADP release primes a plant NLR complex | | Descriptor: | Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, Protein kinase superfamily protein, ... | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Ligand-triggered allosteric ADP release primes a plant NLR complex.
Science, 364, 2019
|
|
7BT6
 
 | |
7BYR
 
 | | BD23-Fab in complex with the S ectodomain trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab23-Fab-Heavy Chain, ... | | Authors: | Zhu, Q, Wang, G, Xiao, J. | | Deposit date: | 2020-04-24 | | Release date: | 2020-06-10 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells.
Cell, 182, 2020
|
|
