3UFB
 
 | | Crystal structure of a modification subunit of a putative type I restriction enzyme from Vibrio vulnificus YJ016 | | Descriptor: | Type I restriction-modification system methyltransferase subunit | | Authors: | Park, S.Y, Lee, H.J, Sun, J, Nishi, K, Song, J.M, Kim, J.S. | | Deposit date: | 2011-11-01 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of a modification subunit of a putative type I restriction enzyme from Vibrio vulnificus YJ016
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2F8L
 
 | |
2AR0
 
 | |
2OKC
 
 | |
3S1S
 
 | | Characterization and crystal structure of the type IIG restriction endonuclease BpuSI | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Shen, B.W, Xu, D, Chan, S.-H, Zheng, Y, Zhu, Y, Xu, S.-Y, Stoddard, B.L. | | Deposit date: | 2011-05-16 | | Release date: | 2011-07-13 | | Last modified: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization and crystal structure of the type IIG restriction endonuclease RM.BpuSI.
Nucleic Acids Res., 39, 2011
|
|
8FS1
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11a (YD905) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{2-[N'-(cyclohexylmethyl)carbamimidamido]ethyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
8FS2
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11b (YD907) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{3-[N'-(cyclohexylmethyl)carbamimidamido]propyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
3KHK
 
 | |
3LKD
 
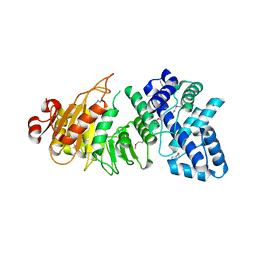 | | Crystal Structure of the type I restriction-modification system methyltransferase subunit from Streptococcus thermophilus, Northeast Structural Genomics Consortium Target SuR80 | | Descriptor: | Type I restriction-modification system methyltransferase subunit | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Mao, M, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Northeast Structural Genomics Consortium Target SuR80
To be published
|
|
7LVV
 
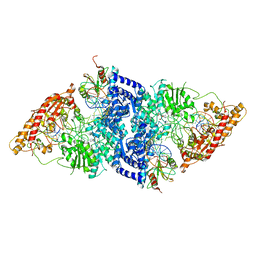 | | cryoEM structure DrdV-DNA complex | | Descriptor: | CALCIUM ION, DNA (27-MER), DNA (28-MER), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Coordination of phage genome degradation versus host genome protection by a bifunctional restriction-modification enzyme visualized by CryoEM.
Structure, 29, 2021
|
|
7LO5
 
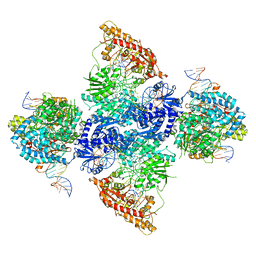 | | cryoEM structure DrdV-DNA complex | | Descriptor: | CALCIUM ION, DNA (27-MER), DNA (28-MER), ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2021-02-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Coordination of phage genome degradation versus host genome protection by a bifunctional restriction-modification enzyme visualized by CryoEM.
Structure, 29, 2021
|
|
7LNI
 
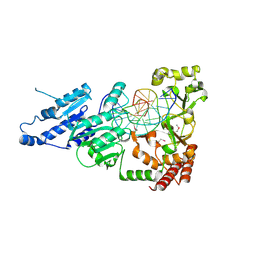 | | SeMet CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-05-19 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix.
Nat Commun, 12, 2021
|
|
7LT5
 
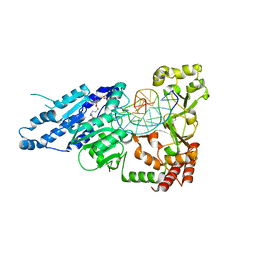 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Cofactor SAH | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix.
Nat Commun, 12, 2021
|
|
7LNJ
 
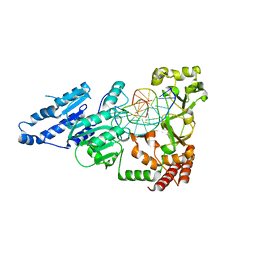 | |
7QW6
 
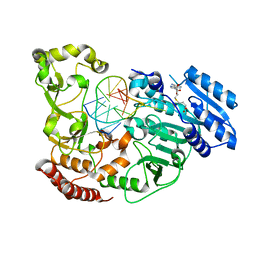 | | Adenine-specific DNA methyltransferase M.BseCI complexed with AdoHcy and cognate hemimethylated DNA duplex | | Descriptor: | Hemimethylated DNA duplex, Modification methylase BseCI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
7QW5
 
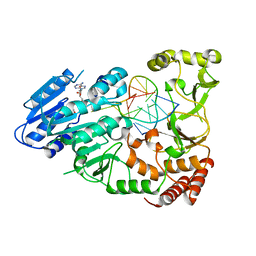 | | Adenine-specific DNA methyltransferase M.BseCI complexed with AdoHcy and cognate unmethylated DNA duplex | | Descriptor: | Modification methylase BseCI, S-ADENOSYL-L-HOMOCYSTEINE, Unmethylated DNA duplex | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
7QW8
 
 | | Adenine-specific DNA methyltransferase M.BseCI | | Descriptor: | Modification methylase BseCI | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
7QW7
 
 | | Adenine-specific DNA methyltransferase M.BseCI complexed with AdoHcy and cognate fully methylated DNA duplex | | Descriptor: | Fully methylated DNA duplex, Modification methylase BseCI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
7RFN
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor SGC8158 | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFM
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor EPZ004777 | | Descriptor: | 1,2-ETHANEDIOL, 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFK
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor Sinefungin | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
7RFL
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor SGC0946 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA Strand 1, ... | | Authors: | Horton, J.R, Cheng, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Repurposing epigenetic inhibitors to target the Clostridioides difficile- specific DNA adenine methyltransferase and sporulation regulator CamA.
Epigenetics, 17, 2022
|
|
8CXV
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 3 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXS
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DNA Strand 1, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXX
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 6 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
