6LCJ
 
 | |
6LCL
 
 | |
6LCK
 
 | |
9AWT
 
 | |
9AYU
 
 | |
9AY8
 
 | |
3MI6
 
 | | Crystal structure of the alpha-galactosidase from Lactobacillus brevis, Northeast Structural Genomics Consortium Target LbR11. | | Descriptor: | Alpha-galactosidase | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Belote, R, Sahdev, S, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-09 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of the alpha-galactosidase from Lactobacillus brevis.
To be Published
|
|
2YFN
 
 | | galactosidase domain of alpha-galactosidase-sucrose kinase, AgaSK | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GALACTOSIDASE-SUCROSE KINASE AGASK, GLYCEROL, ... | | Authors: | Sulzenbacher, G, Bruel, L, Tison-Cervera, M, Pujol, A, Nicoletti, C, Perrier, J, Galinier, A, Ropartz, D, Fons, M, Pompeo, F, Giardina, T. | | Deposit date: | 2011-04-07 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Agask, a Bifunctional Enzyme from the Human Microbiome Coupling Galactosidase and Kinase Activities
J.Biol.Chem., 286, 2011
|
|
6JHP
 
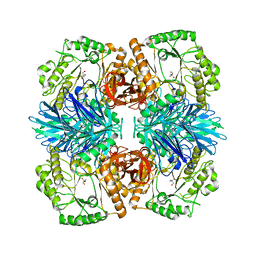 | |
1ZY9
 
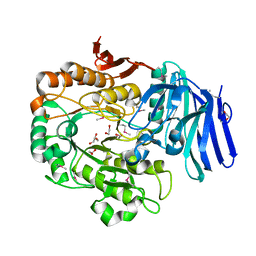 | |
8K7U
 
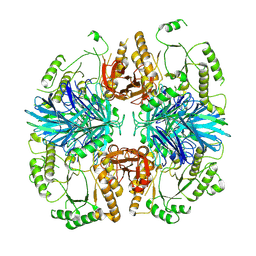 | | the alpha-galactosidase 5 with Cacl2 | | Descriptor: | Alpha-galactosidase | | Authors: | Li, Y.W, Ru, Y.X. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Activity-based metaproteomics driven discovery and enzymological characterization of potential alpha-galactosidases in the mouse gut microbiome.
Commun Chem, 7, 2024
|
|
8K7V
 
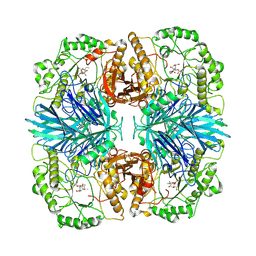 | | the alpha-galactosidase 5 with inhibitor ABP2 | | Descriptor: | 8-azido-1-((1S,2S,3S,4S,5R,6S)-2,3,4-trihydroxy-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptan-7-yl)octan-1-one, Alpha-galactosidase | | Authors: | Li, Y.W, Ru, Y.X. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Activity-based metaproteomics driven discovery and enzymological characterization of potential alpha-galactosidases in the mouse gut microbiome.
Commun Chem, 7, 2024
|
|
8K1A
 
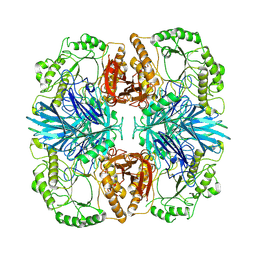 | | the wild-typed alpha-galactosidase 5 | | Descriptor: | Alpha-galactosidase | | Authors: | Li, Y.W, Ru, Y.X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Activity-based metaproteomics driven discovery and enzymological characterization of potential alpha-galactosidases in the mouse gut microbiome.
Commun Chem, 7, 2024
|
|
2XN2
 
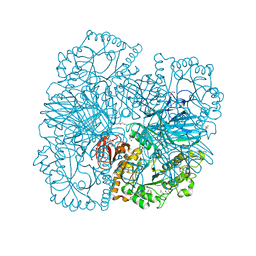 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
2XN1
 
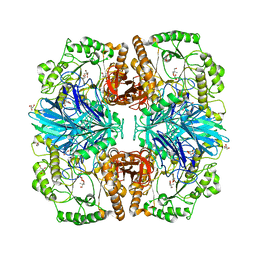 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-GALACTOSIDASE, GLYCEROL | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
5M16
 
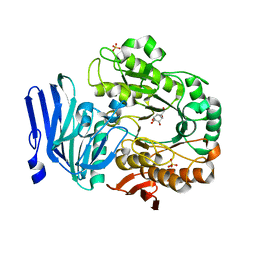 | | Structure of GH36 alpha-galactosidase from Thermotoga maritima in complex with a hydrolysed cyclopropyl carbasugar. | | Descriptor: | (1~{R},2~{S},3~{S},4~{S},5~{S},6~{S})-1-(hydroxymethyl)bicyclo[4.1.0]heptane-2,3,4,5-tetrol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Pengelly, R, Gloster, T. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Snapshots for Mechanism-Based Inactivation of a Glycoside Hydrolase by Cyclopropyl Carbasugars.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5M1I
 
 | | Structure of GH36 alpha-galactosidase from Thermotoga maritima in a covalent complex with a cyclopropyl carbasugar. | | Descriptor: | (1~{R},2~{S},3~{S},4~{S},6~{R})-4-fluoranyl-1-(hydroxymethyl)bicyclo[4.1.0]heptane-2,3-diol, 1,2-ETHANEDIOL, Alpha-galactosidase, ... | | Authors: | Pengelly, R, Gloster, T. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Snapshots for Mechanism-Based Inactivation of a Glycoside Hydrolase by Cyclopropyl Carbasugars.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2XN0
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, PLATINUM (II) ION | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
5M12
 
 | | Structure of GH36 alpha-galactosidase from Thermotoga maritima in complex with intact cyclopropyl-carbasugar. | | Descriptor: | (1~{R},2~{S},3~{S},4~{R},5~{S},6~{S})-5-[3,5-bis(fluoranyl)phenoxy]-1-(hydroxymethyl)bicyclo[4.1.0]heptane-2,3,4-triol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Pengelly, R, Gloster, T. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Snapshots for Mechanism-Based Inactivation of a Glycoside Hydrolase by Cyclopropyl Carbasugars.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5M0X
 
 | |
2YFO
 
 | | GALACTOSIDASE DOMAIN OF ALPHA-GALACTOSIDASE-SUCROSE KINASE, AGASK, in complex with galactose | | Descriptor: | ALPHA-GALACTOSIDASE-SUCROSE KINASE AGASK, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sulzenbacher, G, Bruel, L, Tison-Cervera, M, Pujol, A, Nicoletti, C, Perrier, J, Galinier, A, Ropartz, D, Fons, M, Pompeo, F, Giardina, T. | | Deposit date: | 2011-04-07 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Agask, a Bifunctional Enzyme from the Human Microbiome Coupling Galactosidase and Kinase Activities
J.Biol.Chem., 286, 2011
|
|
6GWG
 
 | | Alpha-galactosidase from Thermotoga maritima in complex with cyclohexene-based carbasugar mimic of galactose covalently linked to the nucleophile | | Descriptor: | (1~{S},2~{S},3~{S})-3-fluoranyl-6-(hydroxymethyl)cyclohex-5-ene-1,2,4-triol, Alpha-galactosidase, GLYCEROL, ... | | Authors: | Gloster, T.M, Oehler, V. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
6GVD
 
 | | Alpha-galactosidase from Thermotoga maritima in complex with cyclohexene-based carbasugar mimic of galactose | | Descriptor: | (1~{S},2~{S},3~{S},4~{S})-5-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Pengelly, R.J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
6GWF
 
 | | Alpha-galactosidase mutant D387A from Thermotoga maritima in complex with intact cyclohexene-based carbasugar mimic of galactose with 2,4-dinitro leaving group | | Descriptor: | (1~{S},2~{S},5~{S},6~{R})-5-(2,4-dinitrophenoxy)-6-fluoranyl-3-(hydroxymethyl)cyclohex-3-ene-1,2-diol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Oehler, V. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
6GTA
 
 | | Alpha-galactosidase mutant D378A from Thermotoga maritima in complex with intact cyclohexene-based carbasugar mimic of galactose with 3,5 difluorophenyl leaving group | | Descriptor: | (1~{R},2~{S},3~{S},6~{S})-6-[3,5-bis(fluoranyl)phenoxy]-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Gloster, T.M, Pengelly, R.J. | | Deposit date: | 2018-06-17 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
