8JML
 
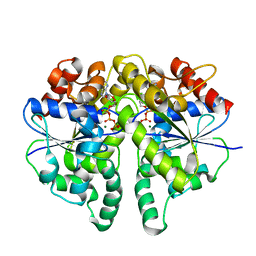 | | Structure of Helicobacter pylori Soj protein mutant, D41A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SpoOJ regulator (Soj) | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
8JMK
 
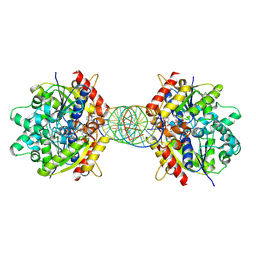 | | Structure of Helicobacter pylori Soj mutant, D41A bound to DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
8JMJ
 
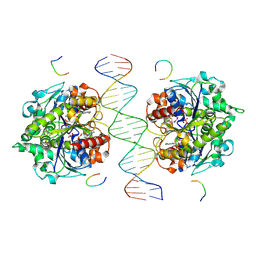 | | Structure of Helicobacter pylori Soj-DNA-Spo0J complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
6RIQ
 
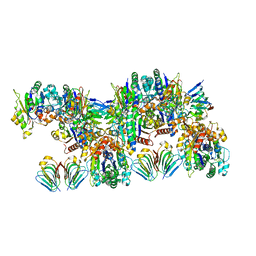 | | MinCD filament from Pseudomonas aeruginosa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MinC, ... | | Authors: | Szewczak-Harris, A, Wagstaff, J, Lowe, J. | | Deposit date: | 2019-04-24 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the MinCD copolymeric filament from Pseudomonas aeruginosa at 3.1 angstrom resolution.
Febs Lett., 593, 2019
|
|
6IUB
 
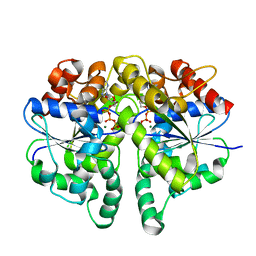 | | Structure of Helicobacter pylori Soj protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SpoOJ regulator (Soj) | | Authors: | Chu, C.H, Yen, C.Y, Sun, Y.J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structures of HpSoj-DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation.
Nucleic Acids Res., 47, 2019
|
|
6IUD
 
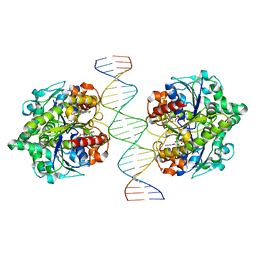 | | Structure of Helicobacter pylori Soj-ADP complex bound to DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Yen, C.Y, Chu, C.H, Sun, Y.J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal structures of HpSoj-DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation.
Nucleic Acids Res., 47, 2019
|
|
6IUC
 
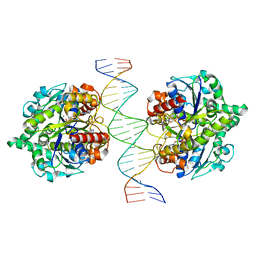 | | Structure of Helicobacter pylori Soj-ATP complex bound to DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Chu, C.H, Yen, C.Y, Sun, Y.J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of HpSoj-DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation.
Nucleic Acids Res., 47, 2019
|
|
4V02
 
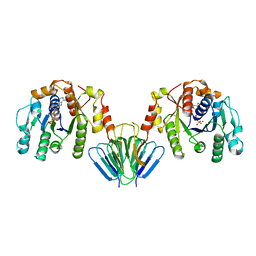 | | MinC:MinD cell division protein complex, Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROBABLE SEPTUM SITE-DETERMINING PROTEIN MINC, ... | | Authors: | Ghosal, D, Lowe, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mincd Cell Division Proteins Form Alternating Copolymeric Cytomotive Filaments.
Nat.Commun., 5, 2014
|
|
4V03
 
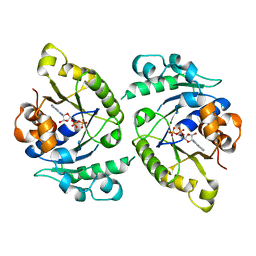 | | MinD cell division protein, Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SITE-DETERMINING PROTEIN | | Authors: | Trambaiolo, D, Lowe, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mincd Cell Division Proteins Form Alternating Copolymeric Cytomotive Filaments.
Nat.Commun., 5, 2014
|
|
4E07
 
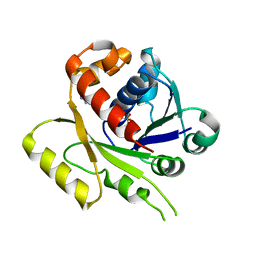 | | ParF-AMPPCP-C2221 form | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4E09
 
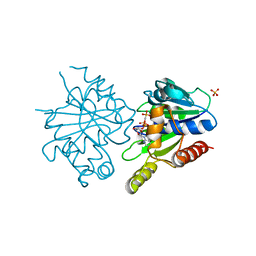 | | Structure of ParF-AMPPCP, I422 form | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF, SULFATE ION | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4E03
 
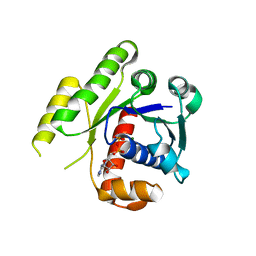 | | Structure of ParF-ADP form 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4DZZ
 
 | | Structure of ParF-ADP, crystal form 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-01 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
3R9J
 
 | | 4.3A resolution structure of a MinD-MinE(I24N) protein complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
3R9I
 
 | | 2.6A resolution structure of MinD complexed with MinE (12-31) peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
3Q9L
 
 | | The structure of the dimeric E.coli MinD-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Septum site-determining protein minD | | Authors: | Wu, W, Park, K.-T, Lutkenhaus, J, Holyoak, T. | | Deposit date: | 2011-01-08 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Determination of the structure of the MinD-ATP complex reveals the orientation of MinD on the membrane and the relative location of the binding sites for MinE and MinC.
Mol.Microbiol., 79, 2011
|
|
3KJI
 
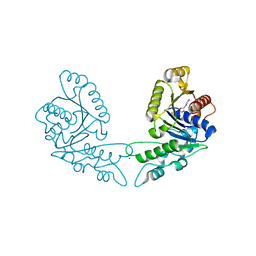 | | Zn and ADP bound state of CooC1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2009-11-03 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of the ATP-Dependent Maturation Factor of Ni,Fe-Containing Carbon Monoxide Dehydrogenases
J.Mol.Biol., 396, 2010
|
|
3KJH
 
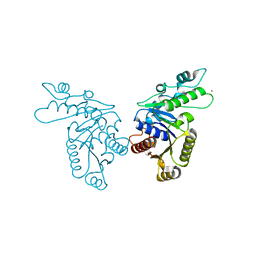 | | Zn-bound state of CooC1 | | Descriptor: | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC, ZINC ION | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2009-11-03 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the ATP-Dependent Maturation Factor of Ni,Fe-Containing Carbon Monoxide Dehydrogenases
J.Mol.Biol., 396, 2010
|
|
3KJG
 
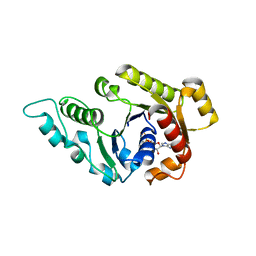 | | ADP-bound state of CooC1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2009-11-03 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the ATP-Dependent Maturation Factor of Ni,Fe-Containing Carbon Monoxide Dehydrogenases
J.Mol.Biol., 396, 2010
|
|
3KJE
 
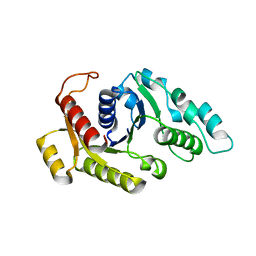 | | Empty state of CooC1 | | Descriptor: | CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2009-11-03 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the ATP-Dependent Maturation Factor of Ni,Fe-Containing Carbon Monoxide Dehydrogenases
J.Mol.Biol., 396, 2010
|
|
3CWQ
 
 | | Crystal structure of chromosome partitioning protein (ParA) in complex with ADP from Synechocystis sp. Northeast Structural Genomics Consortium Target SgR89 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ParA family chromosome partitioning protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, L, Xiao, R, Janjua, H, Maglaqui, M, Foote, E.L, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-04-22 | | Release date: | 2008-05-13 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of chromosome partitioning protein (ParA) in complex with ADP from Synechocystis sp.
To be Published
|
|
1HYQ
 
 | |
1BYI
 
 | | STRUCTURE OF APO-DETHIOBIOTIN SYNTHASE AT 0.97 ANGSTROMS RESOLUTION | | Descriptor: | DETHIOBIOTIN SYNTHASE | | Authors: | Sandalova, T, Schneider, G, Kaeck, H, Lindqvist, Y. | | Deposit date: | 1998-10-15 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of dethiobiotin synthetase at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BS1
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH DETHIOBIOTIN, ADP , INORGANIC PHOSPHATE AND MAGNESIUM | | Descriptor: | 8-AMINO-7-CARBOXYAMINO-NONANOIC ACID WITH ALUMINUM FLUORIDE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kaeck, H, Sandmark, J, Gibson, K.J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1998-08-31 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of two quaternary complexes of dethiobiotin synthetase, enzyme-MgADP-AlF3-diaminopelargonic acid and enzyme-MgADP-dethiobiotin-phosphate; implications for catalysis.
Protein Sci., 7, 1998
|
|
1DAM
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH DETHIOBIOTIN, ADP, INORGANIC PHOSPHATE AND MAGNESIUM | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kaeck, H, Sandmark, J, Gibson, K.J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1998-08-31 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of two quaternary complexes of dethiobiotin synthetase, enzyme-MgADP-AlF3-diaminopelargonic acid and enzyme-MgADP-dethiobiotin-phosphate; implications for catalysis.
Protein Sci., 7, 1998
|
|
