5WRC
 
 | | Crystal structure of proteinase K from Engyodontium album | | Descriptor: | NITRATE ION, PRASEODYMIUM ION, Proteinase K | | Authors: | Sugahara, M, Nakane, T, Suzuki, M, Masuda, T, Inoue, S, Numata, K. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
1ZVJ
 
 | | Structure of Kumamolisin-AS mutant, D164N | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Nakayama, T. | | Deposit date: | 2005-06-02 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Processing, catalytic activity and crystal structures of kumamolisin-As with an engineered active site.
Febs J., 273, 2006
|
|
5MJL
 
 | | Single-shot pink beam serial crystallography: Proteinase K | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.21013784 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
4CFY
 
 | | SAVINASE CRYSTAL STRUCTURES FOR COMBINED SINGLE CRYSTAL DIFFRACTION AND POWDER DIFFRACTION ANALYSIS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN SAVINASE | | Authors: | Frankaer, C.G, Moroz, O.V, Turkenburg, J.P, Aspmo, S.I, Thymark, M, Friis, E.P, Stahla, K, Nielsen, J.E, Wilson, K.S, Harris, P. | | Deposit date: | 2013-11-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Analysis of an Industrial Production Suspension of Bacillus Lentus Subtilisin Crystals by Powder Diffraction: A Powerful Quality-Control Tool.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6F9M
 
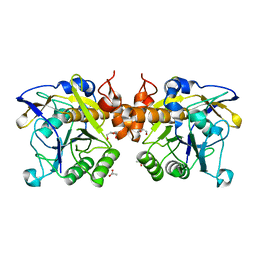 | | The LIPY/F-motif in an intracellular subtilisin protease is involved in inhibition | | Descriptor: | ACETATE ION, SODIUM ION, Serine protease, ... | | Authors: | Bjerga, G.E.K, Larsen, O, Arsin, H, Williamson, A.K, Garcia-Moyano, A, Leiros, I, Puntervoll, P. | | Deposit date: | 2017-12-14 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Mutational analysis of the pro-peptide of a marine intracellular subtilisin protease supports its role in inhibition.
Proteins, 86, 2018
|
|
7JSY
 
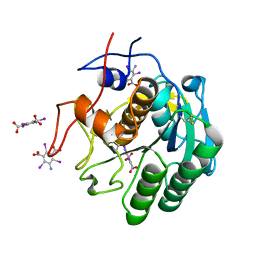 | |
6N4U
 
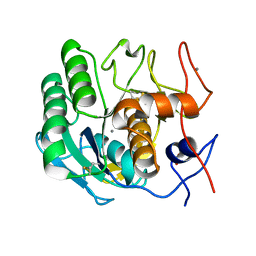 | | MicroED structure of Proteinase K at 2.75A resolution from a single milled crystal. | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2018-11-20 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.75 Å) | | Cite: | Collection of Continuous Rotation MicroED Data from Ion Beam-Milled Crystals of Any Size.
Structure, 27, 2019
|
|
6PKM
 
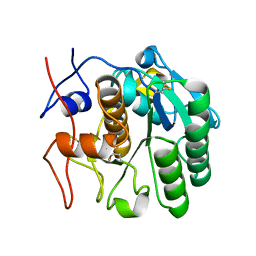 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.17A resolution (#8) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.17 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
5GL8
 
 | |
6PKQ
 
 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.85A resolution (#11) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.85 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
4WOC
 
 | | Proteinase-K Post-Surface Acoustic Waves | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WOB
 
 | | Proteinase-K Pre-Surface Acoustic Wave | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
6O44
 
 | | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Nattokinase, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2019-02-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis.
Biochem. Biophys. Res. Commun., 512, 2019
|
|
5I9S
 
 | | MicroED structure of proteinase K at 1.75 A resolution | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Hattne, J, Shi, D, de la Cruz, M.J, Reyes, F.E, Gonen, T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-06-08 | | Last modified: | 2023-08-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.75 Å) | | Cite: | Modeling truncated pixel values of faint reflections in MicroED images.
J.Appl.Crystallogr., 49, 2016
|
|
1GTJ
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Ala-Phe-cho | | Descriptor: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GTG
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolysin (kscp) | | Descriptor: | CALCIUM ION, KUMAMOLYSIN | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GNS
 
 | | SUBTILISIN BPN' | | Descriptor: | ACETONE, SUBTILISIN BPN' | | Authors: | Almog, O, Gallagher, D.T, Ladner, J.E, Strausberg, S, Alexander, P. | | Deposit date: | 2001-10-06 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Thermostability. Analysis of Stabilizing Mutations in Subtilisin Bpn'.
J.Biol.Chem., 277, 2002
|
|
7Y6M
 
 | |
7C0P
 
 | |
7XR8
 
 | |
5OX2
 
 | |
7S4Z
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Proteinase K | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | Deposit date: | 2021-09-09 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7SKX
 
 | | Ab initio structure of proteinase K from electron-counted MicroED data | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CALCIUM ION, Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Hattne, J, Gonen, T. | | Deposit date: | 2021-10-21 | | Release date: | 2022-06-08 | | Last modified: | 2022-06-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.5 Å) | | Cite: | Ab initio phasing macromolecular structures using electron-counted MicroED data.
Nat.Methods, 19, 2022
|
|
5CW1
 
 | | Proteinase K complexed with 4-iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, IODIDE ION, Proteinase K, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-12-30 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rapid experimental SAD phasing and hot spot identification with halogenated fragments
Iucrj, 3, 2016
|
|
3L1K
 
 | |
