9V2N
 
 | | Macimorelin bound growth hormone secretagogue receptor in complex with Gq | | Descriptor: | Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, R, Sun, J, Liu, H, Guo, S, Zhang, Y, Hu, W, Wang, J, Liu, H, Zhuang, Y, Jiang, Y, Xie, X, Xu, H, Wang, Y. | | Deposit date: | 2025-05-20 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Molecular recognition of two approved drugs Macimorelin and Anamorelin by the growth hormone secretagogue receptor.
Acta Pharmacol.Sin., 2025
|
|
9UY3
 
 | | Anamorelin bound growth hormone secretagogue receptor in complex with Gq | | Descriptor: | 2-azanyl-N-[(2R)-1-[(3S)-3-[dimethylamino(methyl)carbamoyl]-3-(phenylmethyl)piperidin-1-yl]-3-(1H-indol-3-yl)-1-oxidanylidene-propan-2-yl]-2-methyl-propanamide, CHOLESTEROL, Engineered G-alpha-q subunit, ... | | Authors: | Wang, R, Sun, J, Liu, H, Guo, S, Zhang, Y, Hu, W, Wang, J, Liu, H, Zhuang, Y, Jiang, Y, Xie, X, Xu, H.E, Wang, Y. | | Deposit date: | 2025-05-14 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular recognition of two approved drugs Macimorelin and Anamorelin by the growth hormone secretagogue receptor.
Acta Pharmacol.Sin., 2025
|
|
9UWD
 
 | | Cryo-EM structure of inactive-DP1 | | Descriptor: | Prostaglandin D2 receptor,Soluble cytochrome b562 | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2025-05-12 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular basis for ligand recognition and receptor activation of the prostaglandin D2 receptor DP1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9M4T
 
 | | CryoEM structure of the alpha1AAR complex with silodosin | | Descriptor: | Alpha-1A adrenergic receptor, Silodosin | | Authors: | Liu, S.S, Guo, Q, Wang, D.D, Tao, Y.Y, Jiao, H.Z. | | Deposit date: | 2025-03-04 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Molecular mechanism of antagonists recognition and regulation of the alpha 1A - adrenoceptor. ( alpha 1A -Adrenoceptor Antagonist Recognition).
J.Biol.Chem., 2025
|
|
9M4Q
 
 | | CryoEM structure of the alpha1AAR complex with doxazosin | | Descriptor: | Alpha-1A adrenergic receptor, Doxazosin, (S)- | | Authors: | Liu, S.S, Guo, Q, Wang, D.D, Tao, Y.Y, Jiao, H.Z. | | Deposit date: | 2025-03-04 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Molecular mechanism of antagonists recognition and regulation of the alpha 1A - adrenoceptor. ( alpha 1A -Adrenoceptor Antagonist Recognition).
J.Biol.Chem., 2025
|
|
9M1H
 
 | | Cryo-EM structure of PGE2-EP1-Gq complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Meng, X, Xu, Y, Xu, H.E. | | Deposit date: | 2025-02-26 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural Insights into the Activation of Human Prostaglandin E2 Receptor EP1 Subtype by Prostaglandin E2
To Be Published
|
|
9M0D
 
 | | Cryo-EM structure of neurotensin receptor 1 in complex with beta-arrestin 1 | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Sun, D.M, Li, X, Yuan, Q.N, Tian, C.L. | | Deposit date: | 2025-02-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular mechanism of the arrestin-biased agonism of neurotensin receptor 1 by an intracellular allosteric modulator.
Cell Res., 35, 2025
|
|
9LYD
 
 | | Cryo-EM structure of GPR3-1IU9 complex | | Descriptor: | G-protein coupled receptor 3,Aspartate racemase | | Authors: | Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural basis of oligomerization-modulated activation and autoinhibition of orphan receptor GPR3.
Cell Rep, 44, 2025
|
|
9LYB
 
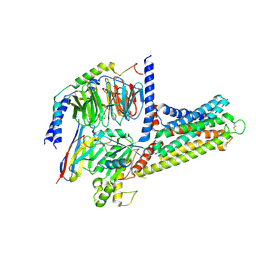 | | Cryo-EM structure of GPR3-G protein-monomer complex | | Descriptor: | G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis of oligomerization-modulated activation and autoinhibition of orphan receptor GPR3.
Cell Rep, 44, 2025
|
|
9LYC
 
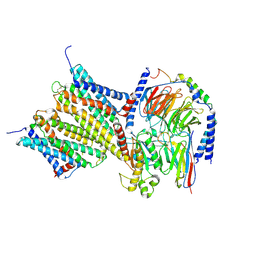 | | Cryo-EM structure of GPR3-G protein-dimer complex | | Descriptor: | G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Hua, T, Liu, Z.J, Li, X.T, Chang, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of oligomerization-modulated activation and autoinhibition of orphan receptor GPR3.
Cell Rep, 44, 2025
|
|
9LRE
 
 | | Cryo-EM structure of the histamine H4 receptor-Gi protein complex (Overall) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptor
To Be Published
|
|
9LRB
 
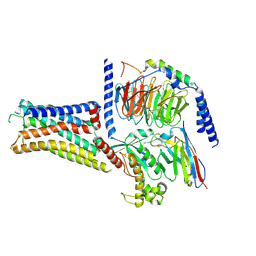 | | Cryo-EM structure of the histamine H1 receptor-Gs protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptors
To Be Published
|
|
9LRD
 
 | | Cryo-EM structure of the histamine H1 receptor-Gi protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptors
To Be Published
|
|
9LRC
 
 | | Cryo-EM structure of the histamine H4 receptor-Gi protein complex (Receptor focused) | | Descriptor: | HISTAMINE, Histamine H4 receptor,Genome polyprotein | | Authors: | Matsuzaki, Y, Sano, F.K, Oshima, H.S, Akasaka, H, Kobayashi, K, Tanaka, T, Itoh, Y, Shihoya, W, Kise, Y, Kusakizako, T, Nureki, O. | | Deposit date: | 2025-01-30 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into ligand recognition and G protein preferences across histamine receptors
To Be Published
|
|
9LMP
 
 | |
9LGM
 
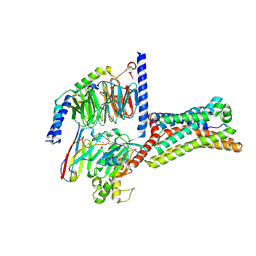 | | Cryo-EM structure of GPR4 complexed with Gs in pH8.0 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2025-01-10 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of stepwise proton sensing-mediated GPCR activation.
Cell Res., 35, 2025
|
|
9MQI
 
 | | Inactive Mu-Opioid Receptor with Nb6M, NabFab, and isoquinuclidine compound #020_E1 | | Descriptor: | Mu-type opioid receptor, NabFab Heavy Chain, NabFab Light Chain, ... | | Authors: | Kim, J.Y, Vigneron, S.F, Billesbolle, C, Manglik, A, Shoichet, B.K. | | Deposit date: | 2025-01-03 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Docking 14 million virtual isoquinuclidines against the mu and kappa opioid receptors reveals dual antagonists-inverse agonists with reduced withdrawal effects.
Biorxiv, 2025
|
|
9MQL
 
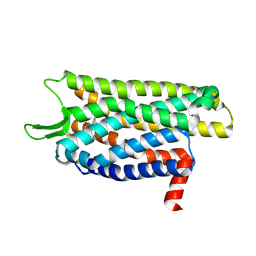 | | Locally-Refined Inactive Kappa-Opioid Receptor with Nb6M, NabFab, and isoquinuclidine compound #020_E1 | | Descriptor: | Kappa-Opioid Receptor, methyl (1S,3R,4S,6S,8M)-2-[(1-ethyl-1H-pyrazol-4-yl)methyl]-8-(3-hydroxyphenyl)-3,4-dimethyl-2-azabicyclo[2.2.2]oct-7-ene-6-carboxylate | | Authors: | Kim, J.Y, Vigneron, S.F, Billesbolle, C, Manglik, A, Shoichet, B.K. | | Deposit date: | 2025-01-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Docking 14 million virtual isoquinuclidines against the mu and kappa opioid receptors reveals dual antagonists-inverse agonists with reduced withdrawal effects.
Biorxiv, 2025
|
|
9MQK
 
 | | Inactive Kappa-Opioid Receptor with Nb6M, NabFab, and isoquinuclidine compound #020_E1 | | Descriptor: | Kappa-Opioid Receptor, NabFab Heavy Chain, NabFab Light Chain, ... | | Authors: | Kim, J.Y, Vigneron, S.F, Billesbolle, C, Manglik, A, Shoichet, B.K. | | Deposit date: | 2025-01-03 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Docking 14 million virtual isoquinuclidines against the mu and kappa opioid receptors reveals dual antagonists-inverse agonists with reduced withdrawal effects.
Biorxiv, 2025
|
|
9MQJ
 
 | | Locally-refined Inactive Mu-Opioid Receptor with Nb6M, NabFab, and isoquinuclidine compound #020_E1 | | Descriptor: | Mu-type opioid receptor, methyl (1S,3R,4S,6S,8M)-2-[(1-ethyl-1H-pyrazol-4-yl)methyl]-8-(3-hydroxyphenyl)-3,4-dimethyl-2-azabicyclo[2.2.2]oct-7-ene-6-carboxylate | | Authors: | Kim, J.Y, Vigneron, S.F, Billesbolle, C, Manglik, A, Shoichet, B.K. | | Deposit date: | 2025-01-03 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Docking 14 million virtual isoquinuclidines against the mu and kappa opioid receptors reveals dual antagonists-inverse agonists with reduced withdrawal effects.
Biorxiv, 2025
|
|
9MQH
 
 | | Inactive Mu-Opioid Receptor with Nb6M, NabFab, and isoquinuclidine compound #33 | | Descriptor: | Mu-type opioid receptor | | Authors: | Kim, J.Y, Vigneron, S.F, Billesbolle, C, Manglik, A, Shoichet, B.K. | | Deposit date: | 2025-01-03 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Docking 14 million virtual isoquinuclidines against the mu and kappa opioid receptors reveals dual antagonists-inverse agonists with reduced withdrawal effects.
Biorxiv, 2025
|
|
9L3Z
 
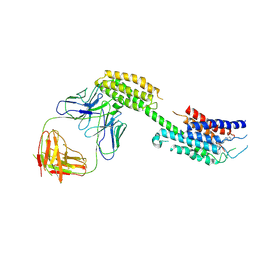 | | Cryo-EM structure of the inactive chemokine-like receptor 1 (CMKLR1) | | Descriptor: | Chemerin-like receptor 1,Soluble cytochrome b562, LRH7-C2, The heavy chain of anti-BRIL Fab, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
9L3W
 
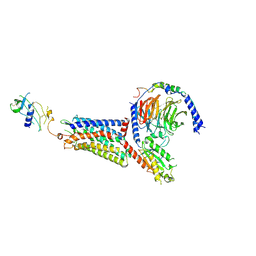 | | Cryo-EM structure of the chemokine-like receptor 1 in complex with chemerin and Gi1 | | Descriptor: | Chemerin-like receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
9L3Y
 
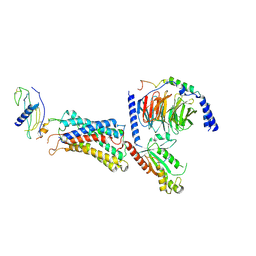 | | Cryo-EM structure of the G-protein coupled receptor 1 (GPR1) in complex with chemerin and Gi1 | | Descriptor: | Chemerin-like receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
9EGO
 
 | | Cannabinoid receptor 1-Gi complex with novel ligand | | Descriptor: | Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tummino, T.A, Iliopoulos-Tsoutsouvas, C, Braz, J.M, O'Brien, E.S, Krishna Kumar, K, Makriyannis, M, Basbaum, A.I, Shoichet, B.K. | | Deposit date: | 2024-11-21 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virtual library docking for cannabinoid-1 receptor agonists with reduced side effects.
Nat Commun, 16, 2025
|
|
