2XY2
 
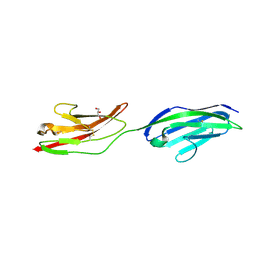 | | CRYSTAL STRUCTURE OF NCAM2 IG1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2YD8
 
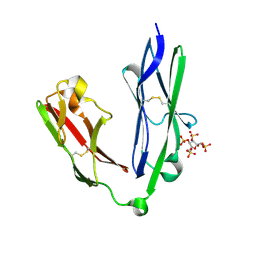 | | Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase LAR in complex with sucrose octasulphate | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE F | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-18 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
2YR3
 
 | | Solution structure of the fourth Ig-like domain from myosin light chain kinase, smooth muscle | | Descriptor: | Myosin light chain kinase, smooth muscle | | Authors: | Qin, X.R, Kurosaki, C, Yoshida, M, Hayahsi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2007-10-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth Ig-like domain from myosin light chain kinase, smooth muscle
To be Published
|
|
2YUZ
 
 | | Solution Structure of 4th Immunoglobulin Domain of Slow Type Myosin-Binding Protein C | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Niraula, T.N, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 4th Immunoglobulin Domain of Slow Type Myosin-Binding Protein C
To be Published
|
|
2YXM
 
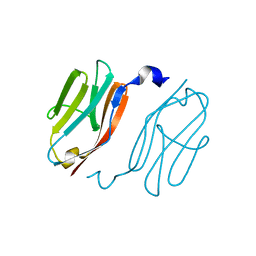 | | Crystal structure of I-set domain of human Myosin Binding ProteinC | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Kishishita, S, Ohsawa, N, Murayama, K, Chen, L, Liu, Z, Terada, T, Shirouzu, M, Wang, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-26 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of I-set domain of human Myosin Binding ProteinC
To be Published
|
|
2YUV
 
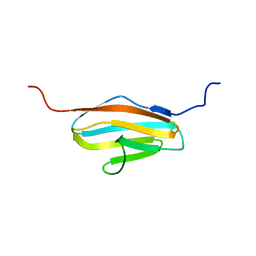 | | Solution Structure of 2nd Immunoglobulin Domain of Slow Type Myosin-Binding Protein C | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Niraula, T.N, Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 2nd Immunoglobulin Domain of Slow Type Myosin-Binding Protein C
To be Published
|
|
2YZ8
 
 | | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556) | | Descriptor: | Obscurin | | Authors: | Saijo, S, Ohsawa, N, Nishino, A, Kishishita, S, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556)
To be Published
|
|
2YD5
 
 | | Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase LAR | | Descriptor: | RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE F | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
2YD1
 
 | | Crystal structure of the N-terminal Ig1-2 module of Drosophila Receptor Protein Tyrosine Phosphatase DLAR | | Descriptor: | GLYCINE, TYROSINE-PROTEIN PHOSPHATASE LAR | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
2Y9R
 
 | |
1BIH
 
 | | CRYSTAL STRUCTURE OF THE INSECT IMMUNE PROTEIN HEMOLIN: A NEW DOMAIN ARRANGEMENT WITH IMPLICATIONS FOR HOMOPHILIC ADHESION | | Descriptor: | HEMOLIN, PHOSPHATE ION | | Authors: | Su, X.-D, Gastinel, L.N, Vaughn, D.E, Faye, I, Poon, P, Bjorkman, P.J. | | Deposit date: | 1998-06-17 | | Release date: | 1998-10-14 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of hemolin: a horseshoe shape with implications for homophilic adhesion.
Science, 281, 1998
|
|
6S9F
 
 | | Drosophila OTK, extracellular domains 3-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tyrosine-protein kinase-like otk | | Authors: | Rozbesky, D, Jones, E.Y. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Drosophila OTK Is a Glycosaminoglycan-Binding Protein with High Conformational Flexibility.
Structure, 28, 2020
|
|
6ZR7
 
 | | X-ray structure of human Dscam Ig7-Ig9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Down syndrome cell adhesion molecule, ... | | Authors: | Kozak, S, Bento, I, Meijers, R. | | Deposit date: | 2020-07-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Homogeneously N-glycosylated proteins derived from the GlycoDelete HEK293 cell line enable diffraction-quality crystallogenesis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5E4I
 
 | | Crystal structure of mouse CNTN5 Ig1-Ig4 domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-5 | | Authors: | Nikolaienko, R.M, Bouyain, S. | | Deposit date: | 2015-10-06 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Interactions Between Contactin Family Members and Protein-tyrosine Phosphatase Receptor Type G in Neural Tissues.
J.Biol.Chem., 291, 2016
|
|
4KC3
 
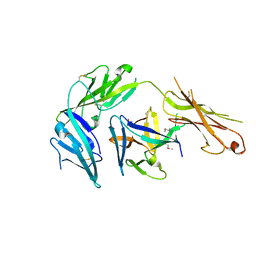 | | Cytokine/receptor binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor-like 1, Interleukin-33 | | Authors: | Liu, X, Wang, X.Q. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2702 Å) | | Cite: | Structural insights into the interaction of IL-33 with its receptors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5NOI
 
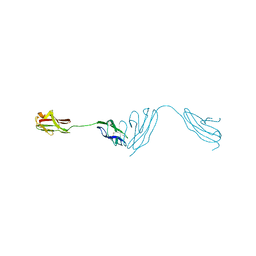 | |
5O5G
 
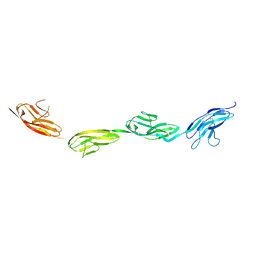 | | Robo1 Ig1 to 4 crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Roundabout homolog 1 | | Authors: | Aleksandrova, N, Gutsche, I, Kandiah, E, Avilov, S.V, Petoukhov, M.V, Seiradake, E, McCarthy, A.A. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Robo1 Forms a Compact Dimer-of-Dimers Assembly.
Structure, 26, 2018
|
|
4RCA
 
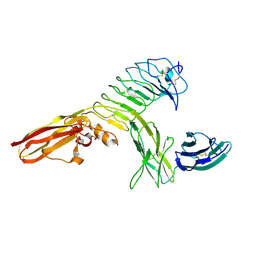 | | Crystal structure of human PTPdelta and human Slitrk1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta, SLIT and NTRK-like protein 1, ... | | Authors: | Kim, H.M, Park, B.S, Kim, D, Lee, S.G. | | Deposit date: | 2014-09-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9908 Å) | | Cite: | Structural basis for LAR-RPTP/Slitrk complex-mediated synaptic adhesion.
Nat Commun, 5, 2014
|
|
5OPE
 
 | | Robo1 Ig1-4 crystals form 2 | | Descriptor: | PHOSPHATE ION, Roundabout homolog 1 | | Authors: | Aleksandrova, N, Gutsche, I, Kandiah, E, Avilov, S.V, Petoukhov, M.V, Seiradake, E, McCarthy, A.A. | | Deposit date: | 2017-08-09 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Robo1 Forms a Compact Dimer-of-Dimers Assembly.
Structure, 26, 2018
|
|
4U7M
 
 | | LRIG1 extracellular domain: Structure and Function Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeats and immunoglobulin-like domains protein 1 | | Authors: | Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | LRIG1 Extracellular Domain: Structure and Function Analysis.
J.Mol.Biol., 427, 2015
|
|
5K6U
 
 | | Sidekick-1 immunoglobulin domains 1-4, crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CESIUM ION, IODIDE ION, ... | | Authors: | Jin, X, Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
3ZYO
 
 | | Crystal structure of the N-terminal leucine rich repeats and immunoglobulin domain of netrin-G ligand-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING PROTEIN 4B, ZINC ION | | Authors: | Seiradake, E, Coles, C.H, Perestenko, P.V, Harlos, K, McIlhinney, R.A.J, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Cell Surface Patterning Through Netring-Ngl Interactions.
Embo J., 30, 2011
|
|
5K6X
 
 | | Sidekick-2 immunoglobulin domains 1-4, crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
6NRR
 
 | | Crystal structure of Dpr11 IG1 bound to DIP-gamma IG+IG2 | | Descriptor: | Defective proboscis extension response 11, isoform B, Dpr-interacting protein gamma, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
5K6P
 
 | | The NMR structure of the m domain tri-helix bundle and C2 of human cardiac Myosin Binding Protein C | | Descriptor: | Myosin-binding protein C, cardiac-type | | Authors: | Michie, K.A, Kwan, A.H, Tung, C.S, Guss, J.M, Trewhella, J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-11-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Highly Conserved Yet Flexible Linker Is Part of a Polymorphic Protein-Binding Domain in Myosin-Binding Protein C.
Structure, 24, 2016
|
|
