2MQH
 
 | |
2YTY
 
 | | Solution structure of the fourth cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
3AQQ
 
 | | Crystal structure of human CRHSP-24 | | Descriptor: | Calcium-regulated heat stable protein 1 | | Authors: | Hou, H, Wang, F, Zhang, W, Wang, D, Li, X, Bartlam, M, Yao, X, Rao, Z. | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | CRHSP-24 is a novel cargo adaptor trafficking between stress granules and processing bodies
To be Published
|
|
2L15
 
 | | Solution Structure of Cold Shock Protein CspA Using Combined NMR and CS-Rosetta method | | Descriptor: | Cold shock protein CspA | | Authors: | Tang, Y, Schneider, W.M, Shen, Y, Raman, S, Inouye, M, Baker, D, Roth, M.J, Montelione, G.T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Fully automated high-quality NMR structure determination of small (2)H-enriched proteins.
J Struct Funct Genomics, 11, 2010
|
|
2ES2
 
 | |
2F52
 
 | |
5YTT
 
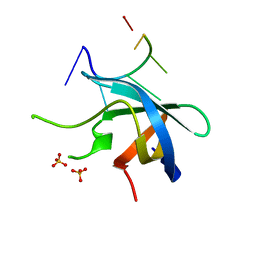 | | Crystal structure of YB1 cold-shock domain in complex with UCAUGU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*UP*GP*U)-3'), SULFATE ION | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTS
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCUUCU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*CP*UP*UP*C)-3'), SULFATE ION | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTV
 
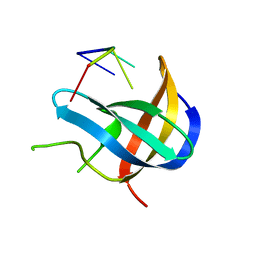 | | Crystal structure of YB1 cold-shock domain in complex with UCAUCU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*UP*CP*U)-3') | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
6A6L
 
 | |
2I5M
 
 | |
2I5L
 
 | |
5XV9
 
 | |
5YTX
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCAACU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*AP*CP*U)-3') | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
6A6J
 
 | | Crystal structure of Zebra fish Y-box protein1 (YB-1) Cold-shock domain in complex with 6mer m5C RNA | | Descriptor: | RNA (5'-R(P*CP*AP*UP*(5MC)P*U)-3'), ZINC ION, Zebra fish Y-box protein1 (YB-1) | | Authors: | Zhang, M.M, Wu, B.X, Huang, Y, Ma, J.B. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay.
Mol.Cell, 75, 2019
|
|
2HAX
 
 | |
2K5N
 
 | | Solution NMR Structure of the N-Terminal Domain of Protein ECA1580 from Erwinia carotovora, Northeast Structural Genomics Consortium Target EwR156A | | Descriptor: | Putative cold-shock protein | | Authors: | Mills, J.L, Eletsky, A, Zhang, Q, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Putative Cold Shock Protein from Erwinia carotovora: Northeast Structural Genomics Consortium Target EwR156a
To be Published
|
|
2KCM
 
 | | Solution NMR structure of the N-terminal OB-domain of SO_1732 from Shewanella oneidensis. Northeast Structural Genomics Consortium Target SoR210A. | | Descriptor: | Cold shock domain family protein | | Authors: | Ramelot, T.A, Maglaqui, M, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-23 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the N-terminal OB-domain of SO_1732 from Shewanella oneidensis. Northeast Structural Genomics Consortium Target SoR210A.
To be Published
|
|
8H1I
 
 | | Crystal structure of PlyGRCS, a bacteriophage Endolysin in complex with Cold shock protein C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cold shock-like protein CspC, ... | | Authors: | Padmanabhan, B, Gopinatha, K, Mandal, M, Saranya, G, Sudhagar, B. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PlyGRCS, a bacteriophage Endolysin in complex with Cold shock protein C
To Be Published
|
|
7OII
 
 | | CspA-70 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
5UDZ
 
 | |
4QQB
 
 | | Structural basis for the assembly of the SXL-UNR translation regulatory complex | | Descriptor: | Protein sex-lethal, Upstream of N-ras, isoform A, ... | | Authors: | Hennig, J, Popowicz, G.M, Sattler, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the assembly of the Sxl-Unr translation regulatory complex.
Nature, 515, 2014
|
|
7ZHR
 
 | |
8OPS
 
 | |
8OPT
 
 | |
