6E4O
 
 | | Structure of apo T. brucei RRM: P4(1)2(1)2 form | | Descriptor: | RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
6E4P
 
 | | Structure of the T. brucei RRM domain in complex with RNA | | Descriptor: | RNA (5'-R(P*UP*UP*UP*U)-3'), RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
6ELD
 
 | |
6F4H
 
 | | Crystal structure of the Drosophila melanogaster SNF/U1-SL2 complex | | Descriptor: | CITRATE ANION, U1 small nuclear ribonucleoprotein A, U1-SL2 | | Authors: | Weber, G, Holton, N, Hall, K.B, DeKoster, G, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
6EZ7
 
 | | Pes4 RRM3 Structure | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein PES4 | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2017-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pes4 RRM3 Structure
To Be Published
|
|
6F4I
 
 | | Crystal structure of Drosophila melanogaster SNF | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Weber, G, Holton, N, Hall, K.B, DeKoster, G, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
6FQ1
 
 | |
6GD2
 
 | | Structure of HuR RRM3 in complex with RNA | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*UP*UP*AP*UP*UP*U)-3') | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6FQR
 
 | |
6GBM
 
 | | Solution structure of FUS-RRM bound to stem-loop RNA | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*GP*AP*UP*UP*AP*CP*AP*AP*UP*UP*CP*UP*AP*UP*UP*UP*GP*CP*C)-3'), RNA-binding protein FUS | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-15 | | Release date: | 2019-02-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
6GD3
 
 | | Structure of HuR RRM3 in complex with RNA (UAUUUA) | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*AP*UP*UP*UP*A)-3'), SODIUM ION | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6G2K
 
 | | Structure of HuR RRM3 in complex with RNA (UUUUUU) | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'), SULFATE ION | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-03-23 | | Release date: | 2018-10-31 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6GC5
 
 | |
6GX6
 
 | | Crystal structure of IMP3 RRM12 in complex with RNA (ACAC) | | Descriptor: | 1,2-ETHANEDIOL, Insulin-like growth factor 2 mRNA-binding protein 3, PHOSPHATE ION, ... | | Authors: | Jia, M, Gut, H, Chao, A.J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of IMP3 RRM12 recognition of RNA.
RNA, 24, 2018
|
|
6HIP
 
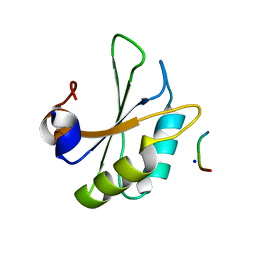 | | Structure of SPF45 UHM bound to HIV-1 Rev ULM | | Descriptor: | HIV-1 Rev (41-49), SODIUM ION, Splicing factor 45, ... | | Authors: | Pabis, M, Corsini, L, Sattler, M. | | Deposit date: | 2018-08-30 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Modulation of HIV-1 gene expression by binding of a ULM motif in the Rev protein to UHM-containing splicing factors.
Nucleic Acids Res., 47, 2019
|
|
8GXC
 
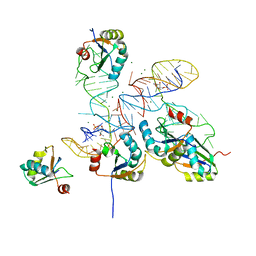 | |
8GXB
 
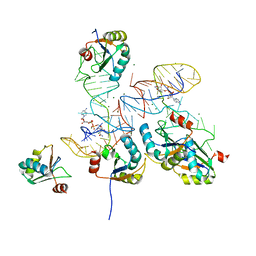 | |
8HNI
 
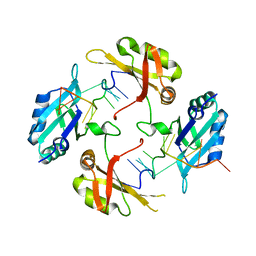 | | hnRNP A2/B1 RRMs in complex with telomeric DNA | | Descriptor: | DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Liu, Y, Abula, A, Xiao, H, Guo, H, Li, T, Zheng, L, Chen, B, Nguyen, H, Ji, X. | | Deposit date: | 2022-12-07 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2023
|
|
1SJ4
 
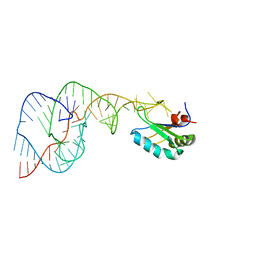 | | Crystal structure of a C75U mutant Hepatitis Delta Virus ribozyme precursor, in Cu2+ solution | | Descriptor: | precursor form of the Hepatitis Delta virus ribozyme, small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H, Doudna, J.A. | | Deposit date: | 2004-03-02 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme
catalysis
Nature, 429, 2004
|
|
2ERR
 
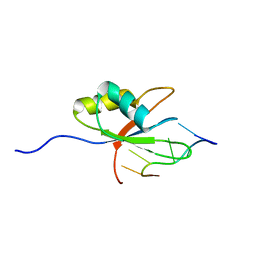 | |
2FC9
 
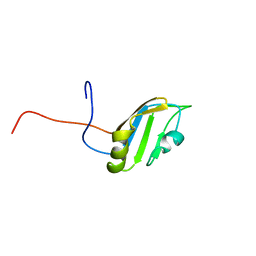 | | Solution structure of the RRM_1 domain of NCL protein | | Descriptor: | NCL protein | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-12 | | Release date: | 2006-06-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM_1 domain of NCL protein
To be published
|
|
2F3J
 
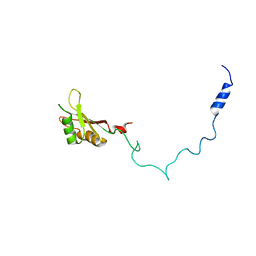 | |
2FC8
 
 | | Solution structure of the RRM_1 domain of NCL protein | | Descriptor: | NCL protein | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-12 | | Release date: | 2006-06-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM_1 domain of NCL protein
To be published
|
|
2F9J
 
 | |
1SXL
 
 | |
