1W4F
 
 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4G
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state folding transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4H
 
 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4I
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4J
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-fast barrier-limited folding in the peripheral subunit-binding domain family.
J. Mol. Biol., 353, 2005
|
|
1W4K
 
 | | Peripheral-subunit binding domains from mesophilic, thermophilic, and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | PYRUVATE DEHYDROGENASE E2 | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Sato, S, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
1W4M
 
 | | Structure of the human pleckstrin DEP domain by multidimensional NMR | | Descriptor: | PLECKSTRIN | | Authors: | Civera, C, Simon, B, Stier, G, Sattler, M, Macias, M.J. | | Deposit date: | 2004-07-26 | | Release date: | 2004-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Human Pleckstrin Dep Domain: Distinct Molecular Features of a Novel Dep Domain Subfamily
Proteins: Struct.,Funct., Genet., 58, 2005
|
|
1W4U
 
 | | NMR solution structure of the ubiquitin conjugating enzyme UbcH5B | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-17 KDA 2 | | Authors: | Houben, K, Dominguez, C, Van Schaik, F.M.A, Timmers, H.T.M, Bonvin, A.M.J.J, Boelens, R. | | Deposit date: | 2004-07-29 | | Release date: | 2004-11-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ubiquitin-Conjugating Enzyme Ubch5B
J.Mol.Biol., 344, 2004
|
|
1W6V
 
 | | Solution structure of the DUSP domain of hUSP15 | | Descriptor: | UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | De Jong, R.D, Ab, E, Diercks, T, Truffault, V, Daniels, M, Kaptein, R, Folkers, G.E. | | Deposit date: | 2004-08-24 | | Release date: | 2006-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Ubiquitin-Specific Protease 15 Dusp Domain.
J.Biol.Chem., 281, 2006
|
|
1W7E
 
 | |
1W86
 
 | |
1W9R
 
 | | Solution Structure of Choline Binding Protein A, Domain R2, the Major Adhesin of Streptococcus pneumoniae | | Descriptor: | CHOLINE BINDING PROTEIN A | | Authors: | Luo, R, Mann, B, Lewis, W.S, Rowe, A, Heath, R, Stewart, M.L, Hamburger, A.E, Bjorkman, P.J, Sivakolundu, S, Lacy, E.R, Tuomanen, E, Kriwacki, R.W. | | Deposit date: | 2004-10-15 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Choline Binding Protein A, the Major Adhesin of Streptococcus Pneumoniae
Embo J., 24, 2005
|
|
1WA7
 
 | | SH3 DOMAIN OF HUMAN LYN TYROSINE KINASE IN COMPLEX WITH A HERPESVIRAL LIGAND | | Descriptor: | HYPOTHETICAL 28.7 KDA PROTEIN IN DHFR 3'REGION (ORF1), TYROSINE-PROTEIN KINASE LYN | | Authors: | Bauer, F, Schweimer, K, Hoffmann, S, Roesch, P, Sticht, H. | | Deposit date: | 2004-10-25 | | Release date: | 2005-07-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigation of the Binding of a Herpesviral Protein to the SH3 Domain of Tyrosine Kinase Lck.
Biochemistry, 41, 2002
|
|
1WA8
 
 | | Solution Structure of the CFP-10.ESAT-6 Complex. Major Virulence Determinants of Pathogenic Mycobacteria | | Descriptor: | 6 KDA EARLY SECRETORY ANTIGENIC TARGET (ESAT-6), ESAT-6 LIKE PROTEIN ESXB | | Authors: | Renshaw, P.S, Lightbody, K.L, Veverka, V, Muskett, F.W, Kelly, G, Frenkiel, T.A, Gordon, S.V, Hewinson, R.G, Burke, B, Norman, J, Williamson, R.A, Carr, M.D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-10-25 | | Release date: | 2005-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Function of the Complex Formed by the Tuberculosis Virulence Factors Cfp-10 and Esat-6
Embo J., 24, 2005
|
|
1WAZ
 
 | |
1WCJ
 
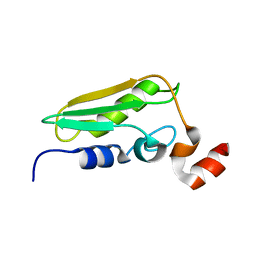 | | Conserved Hypothetical Protein TM0487 from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0487 | | Authors: | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
1WCL
 
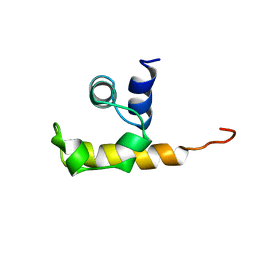 | |
1WCN
 
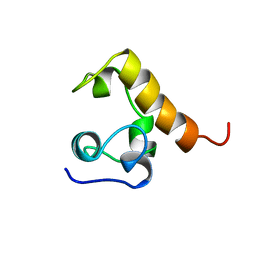 | |
1WCR
 
 | |
1Z2Q
 
 | |
2AGM
 
 | | Solution structure of the R-module from AlgE4 | | Descriptor: | Poly(beta-D-mannuronate) C5 epimerase 4 | | Authors: | Aachmann, F.L, Svanem, B.I, Guntert, P, Petersen, S.B, Valla, S, Wimmer, R. | | Deposit date: | 2005-07-27 | | Release date: | 2006-01-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the R-module: a parallel beta-roll subunit from an Azotobacter vinelandii mannuronan C-5 epimerase.
J.Biol.Chem., 281, 2006
|
|
2AN7
 
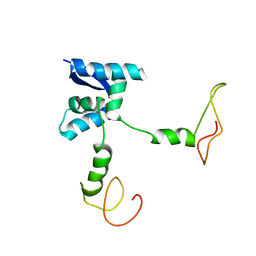 | | Solution structure of the bacterial antidote ParD | | Descriptor: | Protein parD | | Authors: | Oberer, M, Zangger, K, Gruber, K, Keller, W. | | Deposit date: | 2005-08-11 | | Release date: | 2006-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ParD, the antidote of the ParDE toxin antitoxin module, provides the structural basis for DNA and toxin binding.
Protein Sci., 16, 2007
|
|
2AP7
 
 | |
2AVG
 
 | |
2AYX
 
 | | Solution structure of the E.coli RcsC C-terminus (residues 700-949) containing linker region and phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
