8YDP
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 spike protein in complex with Ce9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SARS-CoV-2 inhibiting peptide Ce9, ... | | Authors: | Nakamura, S, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8YDQ
 
 | |
8YDR
 
 | |
8YDS
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant spike protein in complex with Ce59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SARS-CoV-2 inhibiting peptide Ce59, ... | | Authors: | Nakamura, S, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8YDT
 
 | |
8YDU
 
 | |
8YDV
 
 | |
8YDW
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron XBB.1.5 variant spike protein in complex with CeSPIACE | | Descriptor: | GLYCEROL, SARS-CoV-2 inhibiting peptide CeSPIACE, SODIUM ION, ... | | Authors: | Nakamura, S, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8YDX
 
 | | Cryo-EM structure of SARS-CoV-2 spike ectodomain (HexaPro, Omicron BA.2 variant) in complex with CeSPIACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CeSPIACE, Spike glycoprotein | | Authors: | Suzuki, H, Nakamura, S, Fujiyoshi, Y. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8YDY
 
 | | Cryo-EM structure of SARS-CoV-2 spike ectodomain (HexaPro, Omicron BA.5 variant) in complex with CeSPIACE, class 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CeSPIACE, Spike glycoprotein | | Authors: | Suzuki, H, Nakamura, S, Fujiyoshi, Y. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8YDZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike ectodomain (HexaPro, Omicron BA.5 variant) in complex with CeSPIACE, class 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CeSPIACE, Spike glycoprotein | | Authors: | Suzuki, H, Nakamura, S, Fujiyoshi, Y. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8YTS
 
 | | The structure of the cytochrome c546/556 from Thioalkalivibrio paradoxus with unusual UV-Vis spectral features at atomic resolution | | Descriptor: | Cytochrome C, HEME C | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-26 | | Release date: | 2024-04-24 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Relationship between the structure and physicochemical properties of cytochrome c 546/556 from the bacterium Thioalkalivibrio paradoxus ARh1.
Biochem.Biophys.Res.Commun., 778, 2025
|
|
8ZLB
 
 | | EB bound state of hPAC | | Descriptor: | 4-azanyl-6-[[4-[4-[(~{E})-(8-azanyl-1-oxidanyl-5,7-disulfo-naphthalen-2-yl)diazenyl]-3-methyl-phenyl]-2-methyl-phenyl]diazenyl]-5-oxidanyl-naphthalene-1,3-disulfonic acid, Proton-activated chloride channel | | Authors: | Su, N, Chen, X. | | Deposit date: | 2024-05-18 | | Release date: | 2025-07-23 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Discovery and structural basis of endogenous and exogenous inhibitors of the proton-activated-chloride channel.
Cell Rep, 2025
|
|
8ZLL
 
 | | The apo state of hPAC with endogenous cholesterol | | Descriptor: | CHOLESTEROL, Proton-activated chloride channel | | Authors: | Zhang, H, Chen, X. | | Deposit date: | 2024-05-20 | | Release date: | 2025-07-23 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Discovery and structural basis of endogenous and exogenous inhibitors of the proton-activated-chloride channel.
Cell Rep, 2025
|
|
9AZ0
 
 | | Crystal crystal of FC2591 peroxidase from Frankia casuarinae | | Descriptor: | Dyp-type peroxidase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Stogios, P.J, Skarina, T, Choolaei, Z, Yakunin, A, Savchenko, A. | | Deposit date: | 2024-03-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Insights into Lignin-Oxidizing Activity of Bacterial Peroxidases against Soluble Substrates and Kraft Lignin.
Acs Chem.Biol., 20, 2025
|
|
9AZ1
 
 | | Crystal structure of PR9465 peroxidase from Pseudomonas rhizosphaerae | | Descriptor: | CHLORIDE ION, Deferrochelatase, OXYGEN MOLECULE, ... | | Authors: | Stogios, P.J, Skarina, T, Choolaei, Z, Yakunin, A, Savchenko, A. | | Deposit date: | 2024-03-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Insights into Lignin-Oxidizing Activity of Bacterial Peroxidases against Soluble Substrates and Kraft Lignin.
Acs Chem.Biol., 20, 2025
|
|
9AZ2
 
 | | Crystal structure of PF3257 peroxidase from Pseudomonas fluorescens | | Descriptor: | Deferrochelatase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Stogios, P.J, Skarina, T, Choolaei, Z, Yakunin, A, Savchenko, A. | | Deposit date: | 2024-03-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural and Biochemical Insights into Lignin-Oxidizing Activity of Bacterial Peroxidases against Soluble Substrates and Kraft Lignin.
Acs Chem.Biol., 20, 2025
|
|
9B18
 
 | |
9B19
 
 | |
9B1A
 
 | |
9B1B
 
 | | EV-D68 in complex with inhibitor Jun11-78-7 | | Descriptor: | 4-[(propan-2-yl)oxy]-N-{(4M)-4-[1-(2,2,2-trifluoroethyl)-1H-pyrazol-4-yl]quinolin-8-yl}benzamide, Capsid protein VP1, Capsid protein VP4, ... | | Authors: | Klose, T, Kuhn, R.J, Jun, W, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-03-13 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Design of enterovirus D68 capsid inhibitors with in vivo antiviral efficacy
To Be Published
|
|
9B62
 
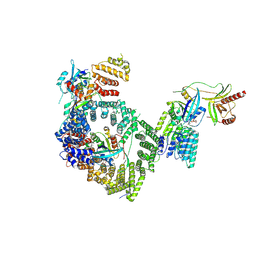 | | Human RANBP2/RAN(GTP)/RANGAP1-SUMO1/UBC9/CRM1/RAN(GTP) - composite map and model | | Descriptor: | E3 SUMO-protein ligase RanBP2, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Lima, C.D, DiMattia, M.A. | | Deposit date: | 2024-03-23 | | Release date: | 2024-10-16 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for a nucleoporin exportin complex between RanBP2, SUMO1-RanGAP1, the E2 Ubc9, Crm1 and the Ran GTPase.
Nat Commun, 16, 2025
|
|
9BS1
 
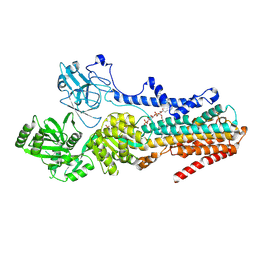 | | Cryo-EM structure of the S. cerevisiae lipid flippase Neo1 bound with PI4P in the E2P state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl di[(9Z)-octadec-9-enoate], BERYLLIUM TRIFLUORIDE ION, Phospholipid-transporting ATPase NEO1 | | Authors: | Duan, H.D, Li, H. | | Deposit date: | 2024-05-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | P4-ATPases control phosphoinositide membrane asymmetry and neomycin resistance.
Nat.Cell Biol., 27, 2025
|
|
9BSN
 
 | |
9BWS
 
 | |
