9CUX
 
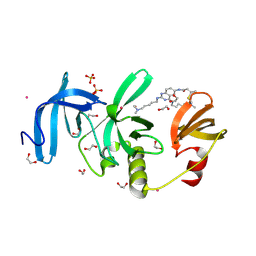 | | Crystal Structure of SETDB1 Tudor domain in complex with UNC100016 | | Descriptor: | (2E)-N-(4-{[6-(dimethylamino)hexyl]amino}-2-{[5-(dimethylamino)pentyl]amino}quinazolin-6-yl)but-2-enamide, 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Silva, M, Dong, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-07-26 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal Structure of SETDB1 Tudor domain in complex with UNC100016
To be published
|
|
9DK4
 
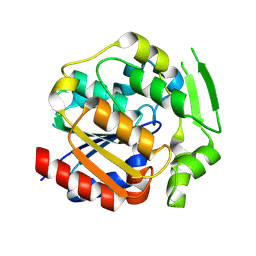 | | ancestral hydroxynitrile lyase with fifteen substitutions | | Descriptor: | Fifteen-substitution variant of an ancestral hydroxynitrile lyase, SODIUM ION | | Authors: | Sarak, S.C, Tan, P, Evans III, R.L, Shi, K, Kazlauskas, R.J, Aihara, H, Pierce, C.T, Walsh, M.E, Greenberg, L.R. | | Deposit date: | 2024-09-07 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | To be published
To Be Published
|
|
9DO4
 
 | |
9DQM
 
 | |
9DR5
 
 | |
9DR6
 
 | |
9DRD
 
 | |
9DRE
 
 | |
9DRF
 
 | |
9F2C
 
 | |
9F56
 
 | |
9F5B
 
 | | Identification of zinc ions in LMO4. | | Descriptor: | LIM domain transcription factor LMO4,LIM domain-binding protein 1, ZINC ION | | Authors: | El Omari, K, Forsyth, I, Mancini, E.J, Wagner, A. | | Deposit date: | 2024-04-28 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Utilizing anomalous signals for element identification in macromolecular crystallography.
Acta Crystallogr D Struct Biol, 2024
|
|
9F5H
 
 | | Crystal structure of MGAT5 bump-and-hole mutant in complex with UDP and M592 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Secreted alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Liu, Y, Bineva-Todd, G, Meek, R, Mazo, L, Piniello, B, Moroz, O.V, Begum, N, Roustan, C, Tomita, S, Kjaer, S, Rovira, C, Davies, G.J, Schumann, B. | | Deposit date: | 2024-04-28 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Bioorthogonal Precision Tool for Human N -Acetylglucosaminyltransferase V.
J.Am.Chem.Soc., 2024
|
|
9F8W
 
 | |
9F9U
 
 | |
9FI9
 
 | | Human PIF + Z48847594 (OCCUPANCY 0.7) | | Descriptor: | ATP-dependent DNA helicase PIF1, MAGNESIUM ION, N-[4-(2-amino-1,3-thiazol-4-yl)phenyl]acetamide, ... | | Authors: | Bax, B.D, Sanders, C.M, Antson, A.A, Brandao-Neto, J. | | Deposit date: | 2024-05-28 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Structure-based discovery of first inhibitors targeting the helicase activity of human PIF1
Nucleic Acids Research, 2024
|
|
9FKX
 
 | | Crystal structure of IL-17A in complex with compound 18 | | Descriptor: | (4~{S})-1-[(~{R})-[2-[(~{S})-[4,4-bis(fluoranyl)cyclohexyl]-[[5-[1,1-bis(fluoranyl)ethyl]-1,3,4-oxadiazol-2-yl]amino]methyl]imidazo[1,2-b]pyridazin-7-yl]-cyclopropyl-methyl]-4-(trifluoromethyl)imidazolidin-2-one, Interleukin-17A | | Authors: | Rondeau, J.M, Lehmann, S, Scheufler, C. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Discovery and In Vivo Exploration of 1,3,4-Oxadiazole and alpha-Fluoroacrylate Containing IL-17 Inhibitors.
J.Med.Chem., 2024
|
|
9FKY
 
 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[4-[(1~{R},3~{R})-3-azanylcyclopentyl]oxyphenyl]-~{N}-[(1~{S})-1-[3-cyano-4-(trifluoromethyl)phenyl]ethyl]propanamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 2024
|
|
9FKZ
 
 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[4-[(1~{R},3~{R})-3-azanylcyclopentyl]oxyphenyl]-~{N}-[(4-chlorophenyl)methyl]prop-2-enamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 2024
|
|
9FL0
 
 | |
9FL3
 
 | | Crystal structure of IL-17A in complex with compound 26 | | Descriptor: | (~{E})-~{N}-[(~{S})-[4,4-bis(fluoranyl)cyclohexyl]-[7-[(1~{S})-2-methoxy-1-[(4~{S})-2-oxidanylidene-4-(trifluoromethyl)imidazolidin-1-yl]ethyl]imidazo[1,2-b]pyridazin-2-yl]methyl]-3-cyclopropyl-2-fluoranyl-prop-2-enamide, Interleukin-17A | | Authors: | Rondeau, J.M, Lehmann, S, Scheufler, C. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Discovery and In Vivo Exploration of 1,3,4-Oxadiazole and alpha-Fluoroacrylate Containing IL-17 Inhibitors.
J.Med.Chem., 2024
|
|
9FMN
 
 | | Structure of Human PADI6 | | Descriptor: | Protein-arginine deiminase type-6 | | Authors: | Mouilleron, S, Walport, L, Williams, J, Marsh, A.J, Hernandez Trapero, R. | | Deposit date: | 2024-06-06 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural insight into the function of human peptidyl arginine deiminase 6.
Comput Struct Biotechnol J, 23, 2024
|
|
9G9Q
 
 | | Crystal structure of PbdA bound to p-methoxybenzoate. | | Descriptor: | 4-METHOXYBENZOIC ACID, Cytochrome P450 CYP199, GLYCEROL, ... | | Authors: | Hinchen, D.J, Wolf, M.E, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Characterization of a cytochrome P450 that catalyzes the O-demethylation of lignin-derived benzoates.
J.Biol.Chem., 2024
|
|
9G9R
 
 | | Crystal structure of PbdA bound to p-ethylbenzoate | | Descriptor: | 4-ethylbenzoic acid, Cytochrome P450 CYP199, GLYCEROL, ... | | Authors: | Hinchen, D.J, Wolf, M.E, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of a cytochrome P450 that catalyzes the O-demethylation of lignin-derived benzoates.
J.Biol.Chem., 2024
|
|
9G9S
 
 | | Crystal structure of PbdA bound to veratrate | | Descriptor: | 3,4-dimethoxybenzoic acid, Cytochrome P450 CYP199, GLYCEROL, ... | | Authors: | Hinchen, D.J, Wolf, M.E, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of a cytochrome P450 that catalyzes the O-demethylation of lignin-derived benzoates.
J.Biol.Chem., 2024
|
|
