1JLA
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLB
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLC
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 | | Descriptor: | HIV-1 RT A-chain, HIV-1 RT B-chain, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLE
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | HIV-1 RT, A-CHAIN, B-CHAIN | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLF
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLG
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JLH
 
 | | Human Glucose-6-phosphate Isomerase | | Descriptor: | phosphoglucose isomerase | | Authors: | Cordeiro, A.T. | | Deposit date: | 2001-07-16 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human phosphoglucose isomerase and analysis of the initial catalytic steps
BIOCHIM.BIOPHYS.ACTA, 1645, 2003
|
|
1JLK
 
 | | Crystal structure of the Mn(2+)-bound form of response regulator Rcp1 | | Descriptor: | MANGANESE (II) ION, Response regulator RCP1, SULFATE ION | | Authors: | Im, Y.J, Rho, S.-H, Park, C.-M, Yang, S.-S, Kang, J.-G, Lee, J.Y, Song, P.-S, Eom, S.H. | | Deposit date: | 2001-07-16 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a cyanobacterial phytochrome response regulator.
Protein Sci., 11, 2002
|
|
1JLL
 
 | | Crystal Structure Analysis of the E197betaA Mutant of E. coli SCS | | Descriptor: | COENZYME A, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Fraser, M.E. | | Deposit date: | 2001-07-16 | | Release date: | 2002-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Two glutamate residues, Glu 208 alpha and Glu 197 beta, are crucial for phosphorylation and dephosphorylation of the active-site histidine residue in succinyl-CoA synthetase.
Biochemistry, 41, 2002
|
|
1JLM
 
 | | I-DOMAIN FROM INTEGRIN CR3, MN2+ BOUND | | Descriptor: | INTEGRIN, MANGANESE (II) ION | | Authors: | Lee, J.-O, Liddington, R. | | Deposit date: | 1996-04-09 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two conformations of the integrin A-domain (I-domain): a pathway for activation?
Structure, 3, 1995
|
|
1JLN
 
 | | Crystal structure of the catalytic domain of protein tyrosine phosphatase PTP-SL/BR7 | | Descriptor: | Protein Tyrosine Phosphatase, receptor type, R | | Authors: | Szedlacsek, S.E, Aricescu, A.R, Fulga, T.A, Renault, L, Scheidig, A.J. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of PTP-SL/PTPBR7 catalytic domain: implications for MAP kinase regulation.
J.Mol.Biol., 311, 2001
|
|
1JLQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 739W94 | | Descriptor: | 2-AMINO-6-(3,5-DIMETHYLPHENYL)SULFONYLBENZONITRILE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 2-Amino-6-arylsulfonylbenzonitriles as non-nucleoside reverse transcriptase inhibitors of HIV-1.
J.Med.Chem., 44, 2001
|
|
1JLR
 
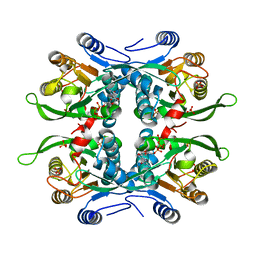 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE GTP COMPLEX 2 MUTANT C128V | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, Uracil Phosphoribosyltransferase | | Authors: | Schumacher, M.A, Bashor, C.J, Otsu, K, Zu, S, Parry, R, Ulmman, B, Brennan, R.G. | | Deposit date: | 2001-07-16 | | Release date: | 2002-01-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JLS
 
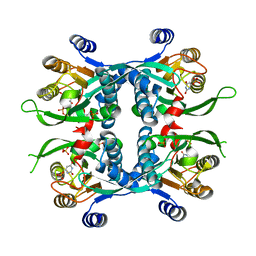 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE URACIL/CPR 2 MUTANT C128V | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Schumacher, M.A, Bashor, C.J, Otsu, K, Zu, S, Parry, R, Ullman, B, Brennan, R.G. | | Deposit date: | 2001-07-16 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JLT
 
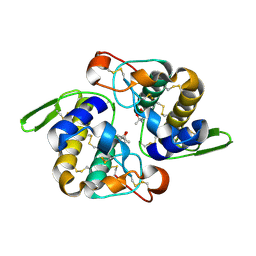 | | Vipoxin Complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHOLIPASE A2, ... | | Authors: | Banumathi, S, Rajashankar, K.R, Notzel, C, Aleksiev, B, Singh, T.P, Genov, N, Betzel, C. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the neurotoxic complex vipoxin at 1.4 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JLV
 
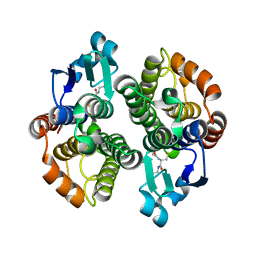 | | Anopheles dirus species B glutathione S-transferases 1-3 | | Descriptor: | GLUTATHIONE, glutathione transferase GST1-3 | | Authors: | Oakley, A.J, Harnnoi, T, Udomsinprasert, R, Jirajaroenrat, K, Ketterman, A.J, Wilce, M.C. | | Deposit date: | 2001-07-16 | | Release date: | 2002-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structures of glutathione S-transferases isozymes 1-3 and 1-4 from Anopheles dirus species B.
Protein Sci., 10, 2001
|
|
1JLW
 
 | | Anopheles dirus species B glutathione S-transferases 1-4 | | Descriptor: | glutathione transferase GST1-4 | | Authors: | Oakley, A.J, Harnnoi, T, Udomsinprasert, R, Jirajaroenrat, K, Ketterman, A.J, Wilce, M.C. | | Deposit date: | 2001-07-16 | | Release date: | 2002-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structures of glutathione S-transferases isozymes 1-3 and 1-4 from Anopheles dirus species B.
Protein Sci., 10, 2001
|
|
1JLX
 
 | | AGGLUTININ IN COMPLEX WITH T-DISACCHARIDE | | Descriptor: | AGGLUTININ, FORMYL GROUP, TOLUENE, ... | | Authors: | Transue, T.R, Smith, A.K, Mo, H, Goldstein, I.J, Saper, M.A. | | Deposit date: | 1997-07-23 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of benzyl T-antigen disaccharide bound to Amaranthus caudatus agglutinin.
Nat.Struct.Biol., 4, 1997
|
|
1JLY
 
 | | CRYSTAL STRUCTURE OF AMARANTHUS CAUDATUS AGGLUTININ | | Descriptor: | AGGLUTININ | | Authors: | Transue, T.R, Smith, A.K, Mo, H, Goldstein, I.J, Saper, M.A. | | Deposit date: | 1997-07-23 | | Release date: | 1997-12-03 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of benzyl T-antigen disaccharide bound to Amaranthus caudatus agglutinin.
Nat.Struct.Biol., 4, 1997
|
|
1JM1
 
 | | Crystal structure of the soluble domain of the Rieske protein II (soxF) from Sulfolobus acidocaldarius | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, Rieske iron-sulfur protein soxF | | Authors: | Boenisch, H, Schmidt, C.L, Schaefer, G, Ladenstein, R. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | The structure of the soluble domain of an archaeal Rieske iron-sulfur protein at 1.1 A resolution.
J.Mol.Biol., 319, 2002
|
|
1JM6
 
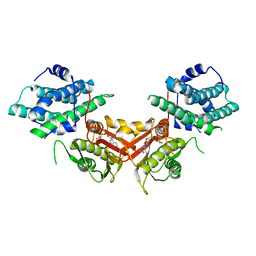 | | Pyruvate dehydrogenase kinase, isozyme 2, containing ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Pyruvate dehydrogenase kinase, ... | | Authors: | Steussy, C.N, Popov, K.M, Bowker-Kinley, M.M, Sloan, R.B, Harris, R.A, Hamilton, J.A. | | Deposit date: | 2001-07-17 | | Release date: | 2001-10-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of pyruvate dehydrogenase kinase. Novel folding pattern for a serine protein kinase.
J.Biol.Chem., 276, 2001
|
|
1JMA
 
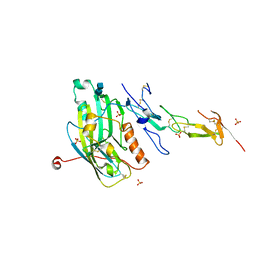 | | CRYSTAL STRUCTURE OF THE HERPES SIMPLEX VIRUS GLYCOPROTEIN D BOUND TO THE CELLULAR RECEPTOR HVEA/HVEM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCOPROTEIN D, HERPESVIRUS ENTRY MEDIATOR, ... | | Authors: | Carfi, A, Willis, S.H, Whitbeck, J.C, Krummenacker, C, Cohen, G.H, Eisenberg, R.J, Wiley, D.C. | | Deposit date: | 2001-07-17 | | Release date: | 2001-09-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Herpes simplex virus glycoprotein D bound to the human receptor HveA.
Mol.Cell, 8, 2001
|
|
1JMC
 
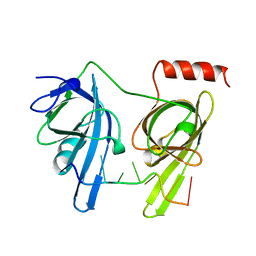 | | SINGLE STRANDED DNA-BINDING DOMAIN OF HUMAN REPLICATION PROTEIN A BOUND TO SINGLE STRANDED DNA, RPA70 SUBUNIT, RESIDUES 183-420 | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*C)-3'), PROTEIN (REPLICATION PROTEIN A (RPA)) | | Authors: | Bochkarev, A, Pfuetzner, R, Edwards, A, Frappier, L. | | Deposit date: | 1996-11-11 | | Release date: | 1997-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA.
Nature, 385, 1997
|
|
1JMF
 
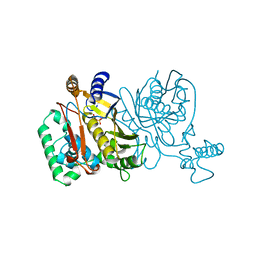 | |
1JMG
 
 | |
