1PH9
 
 | |
1PHJ
 
 | |
1PHN
 
 | |
1PHP
 
 | |
1PHQ
 
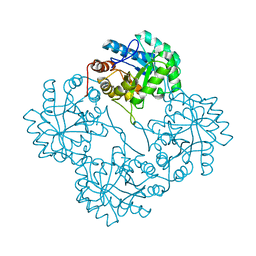 | | Crystal structure of KDO8P synthase in its binary complex with substrate analog E-FPEP | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, 3-FLUORO-2-(PHOSPHONOOXY)PROPANOIC ACID | | Authors: | Vainer, R, Adir, N, Baasov, T, Belakhov, V, Rabkin, E. | | Deposit date: | 2003-05-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic Analysis of the Phosphoenol Pyruvate Binding Site in E. Coli KDO8P Synthase
To be Published
|
|
1PHW
 
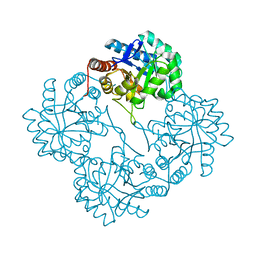 | | Crystal structure of KDO8P synthase in its binary complex with substrate analog 1-deoxy-A5P | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, ANY 5'-MONOPHOSPHATE NUCLEOTIDE | | Authors: | Vainer, R, Belakhov, V, Rabkin, E, Baasov, T, Adir, N. | | Deposit date: | 2003-05-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of Escherichia coli KDO8P synthase complexes reveal the source of catalytic irreversibility.
J.Mol.Biol., 351, 2005
|
|
1PI1
 
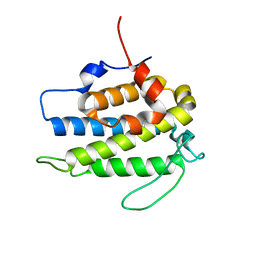 | | Crystal structure of a human Mob1 protein; toward understanding Mob-regulated cell cycle pathways. | | Descriptor: | Mob1A, ZINC ION | | Authors: | Stavridi, E.S, Harris, K.G, Huyen, Y, Bothos, J, Voewerd, P.M, Stayrook, S.E, Jeffrey, P.D, Pavletich, N.P, Luca, F.C. | | Deposit date: | 2003-05-29 | | Release date: | 2003-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a human mob1 protein. Toward understanding mob-regulated cell cycle pathways.
Structure, 11, 2003
|
|
1PI3
 
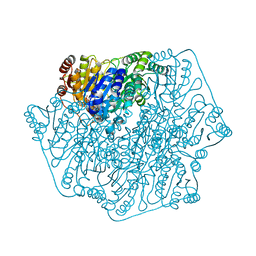 | | E28Q mutant Benzoylformate Decarboxylase From Pseudomonas Putida | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Bera, A.K, Hasson, M.S. | | Deposit date: | 2003-05-29 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High resolution structure of Benzoylformate Decarboxylate E28Q mutant
To be Published
|
|
1PI4
 
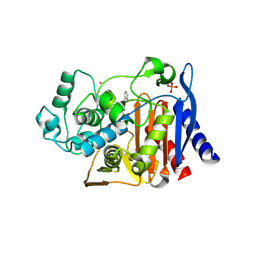 | | Structure of N289A mutant of AmpC in complex with SM3, a phenylglyclboronic acid bearing the cephalothin R1 side chain | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-PHENYLMETHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Roth, T.A, Minasov, G, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2003-05-29 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Thermodynamic cycle analysis and inhibitor design against beta-lactamase.
Biochemistry, 42, 2003
|
|
1PI5
 
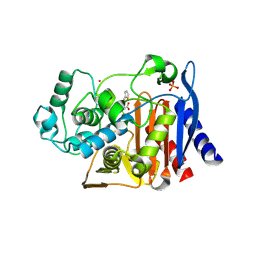 | | Structure of N289A mutant of AmpC in complex with SM2, carboxyphenylglycylboronic acid bearing the cephalothin R1 side chain | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Roth, T.A, Minasov, G, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2003-05-29 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Thermodynamic cycle analysis and inhibitor design against beta-lactamase.
Biochemistry, 42, 2003
|
|
1PIE
 
 | |
1PIN
 
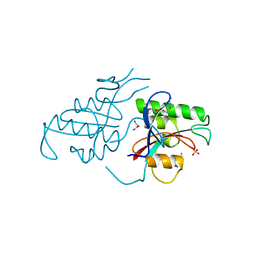 | | PIN1 PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FROM HOMO SAPIENS | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | Authors: | Noel, J.P, Ranganathan, R, Hunter, T. | | Deposit date: | 1998-06-21 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent.
Cell(Cambridge,Mass.), 89, 1997
|
|
1PIW
 
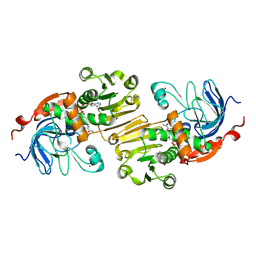 | | APO AND HOLO STRUCTURES OF AN NADP(H)-DEPENDENT CINNAMYL ALCOHOL DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Valencia, E, Larroy, C, Ochoa, W.F, Pares, X, Fita, I, Biosca, J.A. | | Deposit date: | 2003-05-30 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Apo and Holo Structures of an NADP(H)-dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae
J.Mol.Biol., 341, 2004
|
|
1PIX
 
 | | Crystal structure of the carboxyltransferase subunit of the bacterial ion pump glutaconyl-coenzyme A decarboxylase | | Descriptor: | FORMIC ACID, Glutaconyl-CoA decarboxylase A subunit, SULFATE ION | | Authors: | Wendt, K.S, Schall, I, Huber, R, Buckel, W, Jacob, U. | | Deposit date: | 2003-05-30 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the carboxyltransferase subunit of the bacterial sodium ion pump glutaconyl-coenzyme A decarboxylase
Embo J., 22, 2003
|
|
1PJ2
 
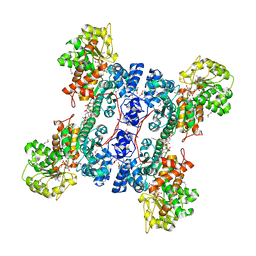 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, cofactor NADH, Mn++, and allosteric activator fumarate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
1PJ3
 
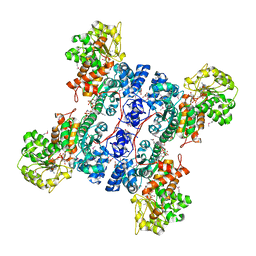 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate pyruvate, cofactor NAD+, Mn++, and allosteric activator fumarate. | | Descriptor: | FUMARIC ACID, MANGANESE (II) ION, NAD-dependent malic enzyme, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
1PJ4
 
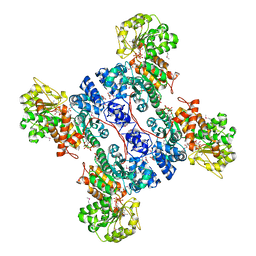 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, ATP, Mn++, and allosteric activator fumarate. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MALATE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-31 | | Release date: | 2003-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
1PJ5
 
 | | Crystal structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with acetate | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, N,N-dimethylglycine oxidase, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
1PJ6
 
 | | Crystal structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with folic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FOLIC ACID, N,N-dimethylglycine oxidase, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
1PJ7
 
 | | Structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with folinic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N,N-dimethylglycine oxidase, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
1PJ9
 
 | | Bacillus circulans strain 251 loop mutant 183-195 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2003-06-02 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improved thermostability of bacillus circulans cyclodextrin glycosyltransferase by the introduction of a salt bridge
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
1PJA
 
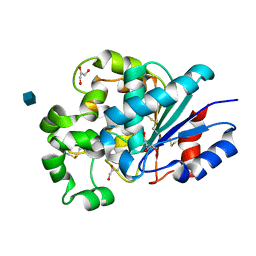 | | The crystal structure of palmitoyl protein thioesterase-2 reveals the basis for divergent substrate specificities of the two lysosomal thioesterases (PPT1 and PPT2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Palmitoyl-protein thioesterase 2 precursor | | Authors: | Calero, G, Gupta, P, Nonato, M.C, Tandel, S, Biehl, E.R, Hofmann, S.L, Clardy, J. | | Deposit date: | 2003-06-02 | | Release date: | 2003-09-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase-2 (PPT2) reveals the basis for divergent substrate specificities of the two lysosomal thioesterases, PPT1 and PPT2.
J.Biol.Chem., 278, 2003
|
|
1PJG
 
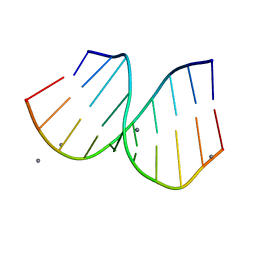 | | RNA/DNA Hybrid Decamer of CAAAGAAAAG/CTTTTCTTTG | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', CALCIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H.
J.Mol.Biol., 334, 2003
|
|
1PJH
 
 | |
1PJI
 
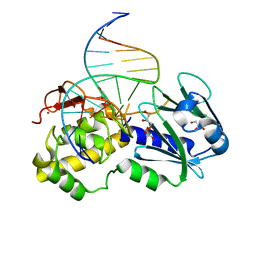 | | Crystal structure of wild type Lactococcus lactis FPG complexed to a 1,3 propanediol containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Pereira, K, Serre, L, Zelwer, C, Castaing, B. | | Deposit date: | 2003-06-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
