4XY6
 
 | |
4XY7
 
 | | Crystal structure of the complex of ribosome inactivating protein from Momordica balsamina with N-acetylglucosamine at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein | | Authors: | Yamini, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-02-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of ribosome inactivating protein from Momordica balsamina with N- acetylglucosamine at 2.5 A resolution
To Be Published
|
|
4XY8
 
 | | Crystal Structure of the bromodomain of BRD9 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, BROMIDE ION, Bromodomain-containing protein 9 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
4XY9
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-(5-bromo-2-methoxyphenyl)-9H-purin-2-amine, Bromodomain-containing protein 4 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
4XYA
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-amine-9H-purine ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-(5-bromo-1-benzofuran-7-yl)-9H-purin-2-amine, Bromodomain-containing protein 4 | | Authors: | Picaud, S, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 9H-Purine Scaffold Reveals Induced-Fit Pocket Plasticity of the BRD9 Bromodomain.
J.Med.Chem., 58, 2015
|
|
4XYB
 
 | | GRANULICELLA M. FORMATE DEHYDROGENASE (FDH) IN COMPLEX WITH NADP(+) AND NaN3 | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, Formate dehydrogenase, ... | | Authors: | Cendron, L, Fogal, S, Beneventi, E, Bergantino, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis for double cofactor specificity in a new formate dehydrogenase from the acidobacterium Granulicella mallensis MP5ACTX8.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4XYC
 
 | | NANOMOLAR INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE 1: SYNTHESIS, BIOLOGICAL EVALUATION AND X-RAY CRYSTALLOGRAPHIC STUDIES | | Descriptor: | 9-phenyl-4H-imidazo[1,2-a]indeno[1,2-e]pyrazin-4-one, Glutamine synthetase 1 | | Authors: | Couturier, C, Silve, S, Morales, R, Ppessegue, B, Llopart, S, Nair, A, Bauer, A, Scheiper, B, poeverlein, c, Ganzhorn, A, Lagrange, S, Bacque, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4XYD
 
 | | Nitric oxide reductase from Roseobacter denitrificans (RdNOR) | | Descriptor: | CALCIUM ION, COPPER (II) ION, FE (III) ION, ... | | Authors: | Crow, A. | | Deposit date: | 2015-02-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the Membrane-intrinsic Nitric Oxide Reductase from Roseobacter denitrificans.
Biochemistry, 55, 2016
|
|
4XYE
 
 | | GRANULICELLA M. FORMATE DEHYDROGENASE (FDH) IN COMPLEX WITH NAD(+) | | Descriptor: | Formate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Cendron, L, Fogal, S, Beneventi, E, Bergantino, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for double cofactor specificity in a new formate dehydrogenase from the acidobacterium Granulicella mallensis MP5ACTX8.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4XYF
 
 | | Crystal structure of c-Met in complex with (S)-5-(8-fluoro-3-(1-(3-(2-methoxyethoxy)quinolin-6-yl)ethyl)-[1,2,4]triazolo[4,3-a]pyridin-6-yl)-3-methylisoxazole | | Descriptor: | 6-{(1S)-1-[8-fluoro-6-(3-methyl-1,2-oxazol-5-yl)[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl}-3-(2-methoxyethoxy)quinoline, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Potent and Selective 8-Fluorotriazolopyridine c-Met Inhibitors.
J.Med.Chem., 58, 2015
|
|
4XYG
 
 | |
4XYH
 
 | |
4XYI
 
 | | Mis16 with H4 peptide | | Descriptor: | Histone H4, Kinetochore protein Mis16 | | Authors: | An, S, Kim, H, Cho, U.-S. | | Deposit date: | 2015-02-02 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mis16 Independently Recognizes Histone H4 and the CENP-ACnp1-Specific Chaperone Scm3sp.
J.Mol.Biol., 427, 2015
|
|
4XYJ
 
 | | Crystal structure of human phosphofructokinase-1 in complex with ATP and Mg, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
4XYK
 
 | | Crystal structure of human phosphofructokinase-1 in complex with ADP, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
4XYL
 
 | | Ca. Korarchaeum cryptofilum ACD1 in complex with coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, alpha subunit of Acyl-CoA synthetase (NDP forming), ... | | Authors: | Weisse, R.H.-J, Scheidig, A.J. | | Deposit date: | 2015-02-02 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of NDP-forming Acetyl-CoA synthetase ACD1 reveals a large rearrangement for phosphoryl transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XYM
 
 | |
4XYN
 
 | | X-ray structure of Ca(2+)-S100B with human RAGE-derived W61 peptide | | Descriptor: | CALCIUM ION, Protein S100-B, Receptor for advanced glycation endproducts-derived peptide (W61) | | Authors: | Jensen, J.L, Indurthi, V.S.K, Neau, D, Vetter, S.W, Colbert, C.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into the binding of the human receptor for advanced glycation end products (RAGE) by S100B, as revealed by an S100B-RAGE-derived peptide complex.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XYO
 
 | | Structure of AgrA LytTR domain | | Descriptor: | 1,2-ETHANEDIOL, Accessory gene regulator A | | Authors: | Gopal, B, Rajasree, K. | | Deposit date: | 2015-02-02 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational features of theStaphylococcus aureusAgrA-promoter interactions rationalize quorum-sensing triggered gene expression.
Biochem Biophys Rep, 6, 2016
|
|
4XYP
 
 | | Crystal structure of a piscine viral fusion protein | | Descriptor: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, CHLORIDE ION, Fusion protein | | Authors: | Cook, J.D, Lee, J.E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Electrostatic Architecture of the Infectious Salmon Anemia Virus (ISAV) Core Fusion Protein Illustrates a Carboxyl-Carboxylate pH Sensor.
J.Biol.Chem., 290, 2015
|
|
4XYQ
 
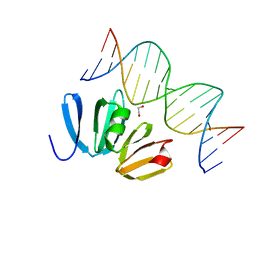 | | Structure of AgrA LytTR domain in complex with promoters | | Descriptor: | 1,2-ETHANEDIOL, Accessory gene regulator A, DNA (5'-D(*AP*AP*TP*AP*CP*TP*TP*AP*AP*CP*TP*GP*TP*TP*AP*A)-3'), ... | | Authors: | Gopal, B, Rajasree, K. | | Deposit date: | 2015-02-03 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational features of theStaphylococcus aureusAgrA-promoter interactions rationalize quorum-sensing triggered gene expression.
Biochem Biophys Rep, 6, 2016
|
|
4XYW
 
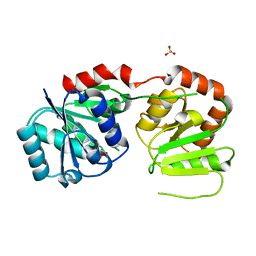 | | Glycosyltransferases WbnH | | Descriptor: | O-antigen biosynthesis glycosyltransferase WbnH, SULFATE ION | | Authors: | Li, F. | | Deposit date: | 2015-02-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Glycosyltransferases WbnH
To Be Published
|
|
4XYX
 
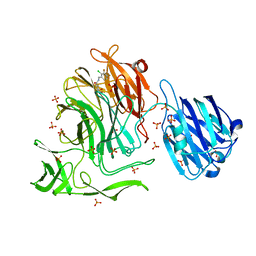 | | NanB plus Optactamide | | Descriptor: | Optactamide, PHOSPHATE ION, Sialidase B | | Authors: | Rogers, G.W, Brear, P, Yang, L, Taylor, G.L, Westwood, N.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Hunt for Serendipitous Allosteric Sites: Discovery of a novel allosteric inhibitor of the bacterial sialidase NanB
To Be Published
|
|
4XYY
 
 | |
4XYZ
 
 | | Crystal structure of K33 linked di-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
