1LVF
 
 | | syntaxin 6 | | Descriptor: | syntaxin 6 | | Authors: | Misura, K.M.S, Bock, J.B, Gonzalez, L.C, Scheller, R.H, Weis, W.I. | | Deposit date: | 2002-05-28 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the amino-terminal domain of syntaxin 6, a SNAP-25 C homolog.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LVG
 
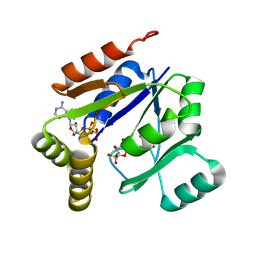 | | Crystal structure of mouse guanylate kinase in complex with GMP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, ... | | Authors: | Sekulic, N, Shuvalova, L, Spangenberg, O, Konrad, M, Lavie, A. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the closed
conformation of mouse guanylate kinase.
J.Biol.Chem., 277, 2002
|
|
1LVH
 
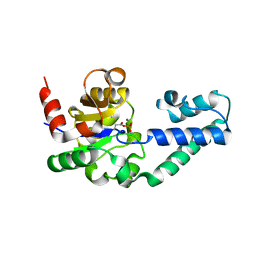 | | The Structure of Phosphorylated beta-phosphoglucomutase from Lactoccocus lactis to 2.3 angstrom resolution | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2002-05-28 | | Release date: | 2002-08-14 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Caught in the act: the structure of phosphorylated beta-phosphoglucomutase from Lactococcus lactis.
Biochemistry, 41, 2002
|
|
1LVL
 
 | |
1LVM
 
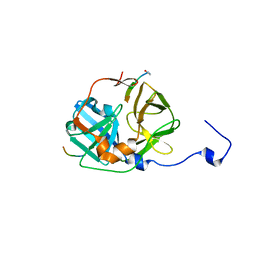 | | CATALYTICALLY ACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH PRODUCT | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
1LVN
 
 | |
1LVW
 
 | | Crystal structure of glucose-1-phosphate thymidylyltransferase, RmlA, complex with dTDP | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Dong, A, Christendat, D, Pai, E.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-05-29 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glucose-1-phosphate thymidylyltransferase, RmlA, complex with dTDP
To be Published
|
|
1LVY
 
 | | PORCINE ELASTASE | | Descriptor: | CALCIUM ION, ELASTASE, SULFATE ION | | Authors: | Schiltz, M, Prange, T. | | Deposit date: | 1996-07-20 | | Release date: | 1997-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | High-pressure krypton gas and statistical heavy-atom refinement: a successful combination of tools for macromolecular structure determination.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1LW0
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LW1
 
 | |
1LW2
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 1051U91 | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LW4
 
 | |
1LW5
 
 | |
1LW6
 
 | |
1LW9
 
 | | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H.M, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-05-30 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability.
Biophys.Chem., 100, 2003
|
|
1LWC
 
 | | CRYSTAL STRUCTURE OF M184V MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LWD
 
 | |
1LWE
 
 | | CRYSTAL STRUCTURE OF M41L/T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMN) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LWG
 
 | | Multiple Methionine Substitutions are Tolerated in T4 Lysozyme and have Coupled Effects on Folding and Stability | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme, ... | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H.M, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.M. | | Deposit date: | 2002-05-31 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability.
Biophys.Chem., 100, 2003
|
|
1LWK
 
 | | Multiple Methionine Substitutions are Tolerated in T4 Lysozyme and have Coupled Effects on Folding and Stability | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H.M, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.M. | | Deposit date: | 2002-05-31 | | Release date: | 2003-05-20 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability.
Biophys.Chem., 100, 2003
|
|
1LWS
 
 | | Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence | | Descriptor: | CALCIUM ION, ENDONUCLEASE PI-SCEI, PI-SceI DNA recognition region bottom strand, ... | | Authors: | Moure, C.M, Gimble, F.S, Quiocho, F.A. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence.
Nat.Struct.Biol., 9, 2002
|
|
1LWT
 
 | |
1LWV
 
 | | Borohydride-trapped hOgg1 Intermediate Structure Co-Crystallized with 8-aminoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-AMINOGUANINE, ... | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
1LWW
 
 | | Borohydride-trapped hOgg1 Intermediate Structure Co-Crystallized with 8-bromoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-BROMOGUANINE, ... | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
