1JK7
 
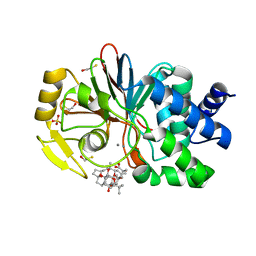 | | CRYSTAL STRUCTURE OF THE TUMOR-PROMOTER OKADAIC ACID BOUND TO PROTEIN PHOSPHATASE-1 | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, OKADAIC ACID, ... | | Authors: | Maynes, J.T, Bateman, K.S, Cherney, M.M, Das, A.K, James, M.N. | | Deposit date: | 2001-07-11 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the tumor-promoter okadaic acid bound to protein phosphatase-1.
J.Biol.Chem., 276, 2001
|
|
1JK8
 
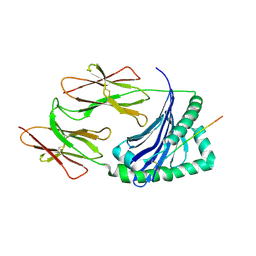 | |
1JK9
 
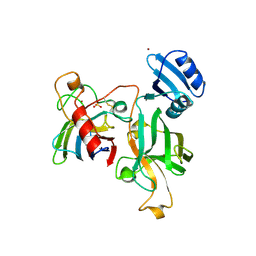 | | Heterodimer between H48F-ySOD1 and yCCS | | Descriptor: | SULFATE ION, ZINC ION, copper chaperone for superoxide dismutase, ... | | Authors: | Lamb, A.L, Torres, A.S, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 2001-07-11 | | Release date: | 2001-09-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Heterodimeric structure of superoxide dismutase in complex with its metallochaperone.
Nat.Struct.Biol., 8, 2001
|
|
1JKA
 
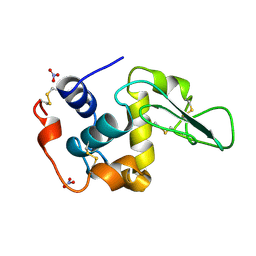 | | HUMAN LYSOZYME MUTANT WITH GLU 35 REPLACED BY ASP | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
1JKB
 
 | | HUMAN LYSOZYME MUTANT WITH GLU 35 REPLACED BY ALA | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
1JKC
 
 | | HUMAN LYSOZYME MUTANT WITH TRP 109 REPLACED BY PHE | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
1JKD
 
 | | HUMAN LYSOZYME MUTANT WITH TRP 109 REPLACED BY ALA | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Muraki, M, Harata, K, Goda, S, Nagahora, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Importance of van der Waals contact between Glu 35 and Trp 109 to the catalytic action of human lysozyme.
Protein Sci., 6, 1997
|
|
1JKG
 
 | | Structural basis for the recognition of a nucleoporin FG-repeat by the NTF2-like domain of TAP-p15 mRNA nuclear export factor | | Descriptor: | TAP, p15 | | Authors: | Fribourg, S, Braun, I.C, Izaurralde, E, Conti, E. | | Deposit date: | 2001-07-12 | | Release date: | 2001-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor.
Mol.Cell, 8, 2001
|
|
1JKH
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-12 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1JKJ
 
 | | E. coli SCS | | Descriptor: | COENZYME A, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Fraser, M.E. | | Deposit date: | 2001-07-12 | | Release date: | 2002-01-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Two glutamate residues, Glu 208 alpha and Glu 197 beta, are crucial for phosphorylation and dephosphorylation of the active-site histidine residue in succinyl-CoA synthetase.
Biochemistry, 41, 2002
|
|
1JKK
 
 | | 2.4A X-RAY STRUCTURE OF TERNARY COMPLEX OF A CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE WITH ATP ANALOGUE AND MG. | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-07-12 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
1JKL
 
 | | 1.6A X-RAY STRUCTURE OF BINARY COMPLEX OF A CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE WITH ATP ANALOGUE | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-07-12 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
1JKO
 
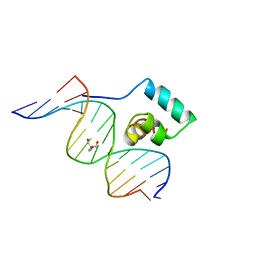 | | Testing the Water-Mediated HIN Recombinase DNA Recognition by Systematic Mutations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*CP*TP*TP*AP*CP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*GP*TP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-12 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1JKP
 
 | | Testing the Water-Mediated HIN Recombinase DNA Recognition by Systematic Mutations | | Descriptor: | 5'-D(*AP*TP*CP*TP*TP*CP*TP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*AP*GP*AP*AP*GP*A)-3', DNA-INVERTASE HIN | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1JKQ
 
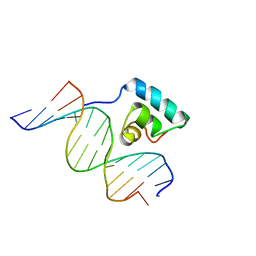 | | Testing the Water-Mediated HIN Recombinase DNA Recognition by Systematic Mutations | | Descriptor: | 5'-D(*AP*TP*CP*TP*TP*AP*TP*AP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3', DNA-INVERTASE HIN | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1JKR
 
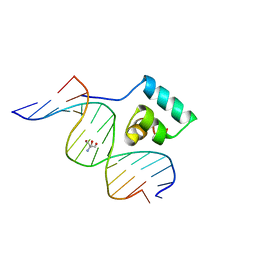 | | Testing the Water-Mediated HIN Recombinase DNA Recognition by Systematic Mutations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*CP*TP*TP*GP*TP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*AP*CP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1JKS
 
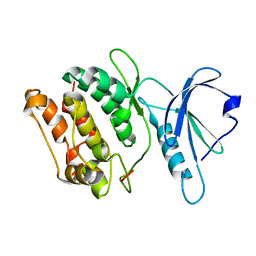 | | 1.5A X-RAY STRUCTURE OF APO FORM OF A CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-07-13 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
1JKT
 
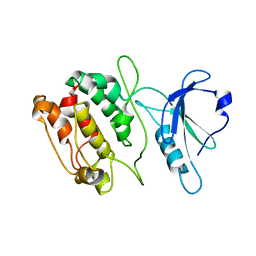 | | TETRAGONAL CRYSTAL FORM OF A CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-07-13 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
1JKX
 
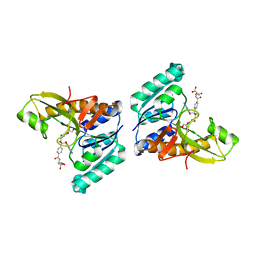 | | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase | | Descriptor: | N-[5'-O-PHOSPHONO-RIBOFURANOSYL]-2-[2-HYDROXY-2-[4-[GLUTAMIC ACID]-N-CARBONYLPHENYL]-3-[2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL]-PROPANYLAMINO]-ACETAMIDE, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Greasley, S.E, Marsilje, T.H, Cai, H, Baker, S, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase.
Biochemistry, 40, 2001
|
|
1JL0
 
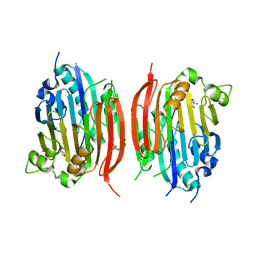 | | Structure of a Human S-Adenosylmethionine Decarboxylase Self-processing Ester Intermediate and Mechanism of Putrescine Stimulation of Processing as Revealed by the H243A Mutant | | Descriptor: | 1,4-DIAMINOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYLMETHIONINE DECARBOXYLASE PROENZYME | | Authors: | Ekstrom, J.L, Tolbert, W.D, Xiong, H, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-07-13 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a human S-adenosylmethionine decarboxylase self-processing ester intermediate and mechanism of putrescine stimulation of processing as revealed by the H243A mutant.
Biochemistry, 40, 2001
|
|
1JL3
 
 | | Crystal Structure of B. subtilis ArsC | | Descriptor: | ARSENATE REDUCTASE, SULFATE ION | | Authors: | Su, X.-D, Bennett, M.S. | | Deposit date: | 2001-07-15 | | Release date: | 2001-10-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JL4
 
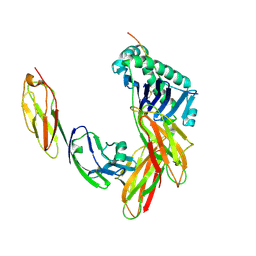 | | CRYSTAL STRUCTURE OF THE HUMAN CD4 N-TERMINAL TWO DOMAIN FRAGMENT COMPLEXED TO A CLASS II MHC MOLECULE | | Descriptor: | H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, A-K ALPHA CHAIN, A-K BETA CHAIN, ... | | Authors: | Wang, J.-H, Meijers, R, Reinherz, E.L. | | Deposit date: | 2001-07-15 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JL5
 
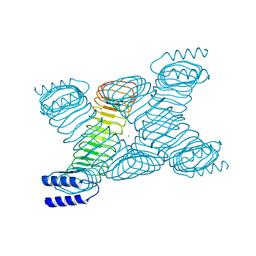 | | Novel Molecular Architecture of YopM-a Leucine-rich Effector Protein from Yersinia pestis | | Descriptor: | CALCIUM ION, outer protein YopM | | Authors: | Evdokimov, A.G, Anderson, D.E, Routzahn, K.M, Waugh, D.S. | | Deposit date: | 2001-07-15 | | Release date: | 2001-10-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit.
J.Mol.Biol., 312, 2001
|
|
1JL8
 
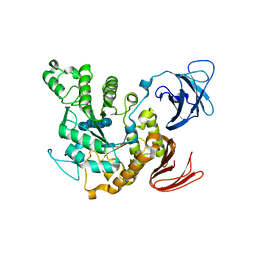 | | Complex of alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with beta-cyclodextrin based on a co-crystallization with methyl beta-cyclodextrin | | Descriptor: | ALPHA-AMYLASE II, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1JL9
 
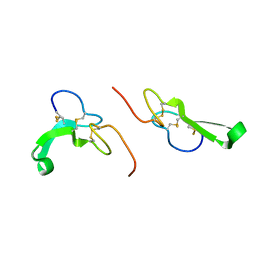 | | Crystal Structure of Human Epidermal Growth Factor | | Descriptor: | EPIDERMAL GROWTH FACTOR | | Authors: | Lu, H.S, Chai, J.J, Li, M, Huang, B.R, He, C.H, Bi, R.C. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human epidermal growth factor and its dimerization
J.Biol.Chem., 276, 2001
|
|
