1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | Authors: | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | Deposit date: | 2000-03-17 | | Release date: | 2000-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1C7Y
 
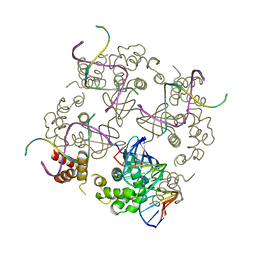 | | E.COLI RUVA-HOLLIDAY JUNCTION COMPLEX | | Descriptor: | DNA (5'-D(P*DAP*DAP*DGP*DTP*DTP*DGP*DGP*DGP*DAP*DTP*DTP*DGP*DT)-3'), DNA (5'-D(P*DCP*DAP*DAP*DTP*DCP*DCP*DCP*DAP*DAP*DCP*DTP*DT)-3'), DNA (5'-D(P*DCP*DGP*DAP*DAP*DTP*DGP*DTP*DGP*DTP*DGP*DTP*DCP*DT)-3'), ... | | Authors: | Ariyoshi, M, Nishino, T, Iwasaki, H, Shinagawa, H, Morikawa, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the holliday junction DNA in complex with a single RuvA tetramer.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C82
 
 | |
1C8B
 
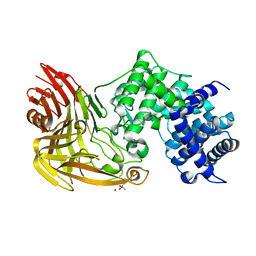
 | | CRYSTAL STRUCTURE OF A NOVEL GERMINATION PROTEASE FROM SPORES OF BACILLUS MEGATERIUM: STRUCTURAL REARRANGEMENTS AND ZYMOGEN ACTIVATION | | Descriptor: | SPORE PROTEASE | | Authors: | Ponnuraj, K, Rowland, S, Nessi, C, Setlow, P, Jedrzejas, M.J. | | Deposit date: | 2000-05-03 | | Release date: | 2001-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a novel germination protease from spores of Bacillus megaterium: structural arrangement and zymogen activation.
J.Mol.Biol., 300, 2000
|
|
1C8C
 
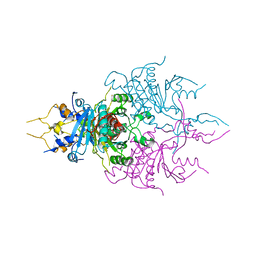
 | | CRYSTAL STRUCTURES OF THE CHROMOSOMAL PROTEINS SSO7D/SAC7D BOUND TO DNA CONTAINING T-G MISMATCHED BASE PAIRS | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3', DNA-BINDING PROTEIN 7A | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Liaw, Y.-C, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 2000-05-04 | | Release date: | 2001-05-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs.
J.Mol.Biol., 303, 2000
|
|
1C8D
 
 | |
1C8E
 
 | |
1C8F
 
 | | FELINE PANLEUKOPENIA VIRUS EMPTY CAPSID STRUCTURE | | Descriptor: | CALCIUM ION, FELINE PANLEUKOPENIA VIRUS CAPSID | | Authors: | Rossmann, M.G, Simpson, A.A. | | Deposit date: | 2000-05-05 | | Release date: | 2000-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses.
J.Mol.Biol., 300, 2000
|
|
1C8G
 
 | | FELINE PANLEUKOPENIA VIRUS EMPTY CAPSID STRUCTURE | | Descriptor: | CALCIUM ION, FELINE PANLEUKOPENIA VIRUS CAPSID | | Authors: | Rossmann, M.G, Simpson, A.A. | | Deposit date: | 2000-05-05 | | Release date: | 2000-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses.
J.Mol.Biol., 300, 2000
|
|
1C8H
 
 | |
1C8N
 
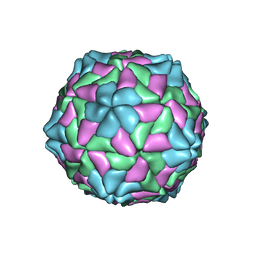 | | TOBACCO NECROSIS VIRUS | | Descriptor: | CALCIUM ION, COAT PROTEIN | | Authors: | Oda, Y, Fukuyama, K. | | Deposit date: | 2000-05-20 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of tobacco necrosis virus at 2.25 A resolution.
J.Mol.Biol., 300, 2000
|
|
1C8Q
 
 | |
1C8T
 
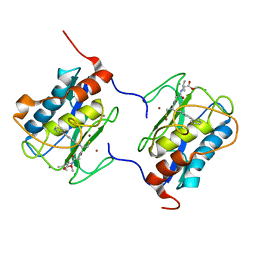 | | HUMAN STROMELYSIN-1 (E202Q) CATALYTIC DOMAIN COMPLEXED WITH RO-26-2812 | | Descriptor: | 2-(2-{2-[(BIPHENYL-4-YLMETHYL)-AMINO]-3-MERCAPTO-PENTANOYLAMINO}-ACETYLAMINO)-3-METHYL-BUTYRIC ACID METHYL ESTER, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Steele, D.L, el-Kabbani, O, Dunten, P, Crowther, R.L. | | Deposit date: | 1999-07-29 | | Release date: | 2000-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expression, characterization and structure determination of an active site mutant (Glu202-Gln) of mini-stromelysin-1.
Protein Eng., 13, 2000
|
|
1C8U
 
 | |
1C8V
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLTHIO)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-07-30 | | Release date: | 2000-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
1C8X
 
 | | Endo-Beta-N-Acetylglucosaminidase H, D130E Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H, PHOSPHATE ION | | Authors: | Rao, V, Tao, C, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C8Y
 
 | | Endo-Beta-N-Acetylglucosaminidase H, D130A Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H, ZINC ION | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C90
 
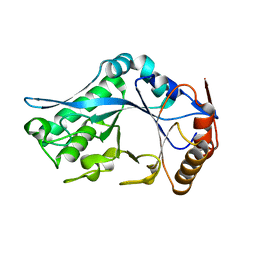 | | Endo-Beta-N-Acetylglucosaminidase H, E132Q Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Tao, C, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Assp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C91
 
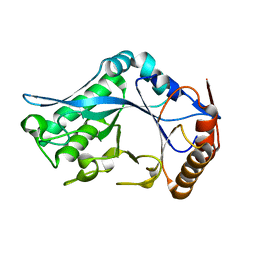 | | Endo-Beta-N-Acetylglucosaminidase H, E132D | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C92
 
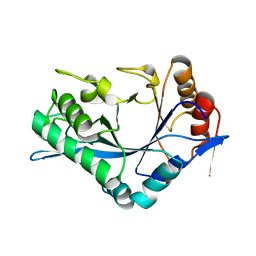 | | Endo-Beta-N-Acetylglucosaminidase H, E132A Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C93
 
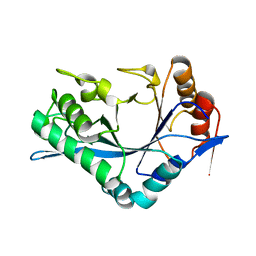 | | Endo-Beta-N-Acetylglucosaminidase H, D130N/E132Q Double Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C9B
 
 | |
1C9D
 
 | |
1C9E
 
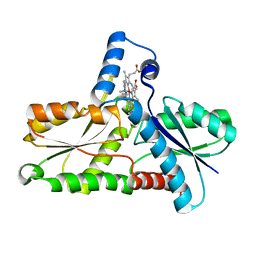 | | STRUCTURE OF FERROCHELATASE WITH COPPER(II) N-METHYLMESOPORPHYRIN COMPLEX BOUND AT THE ACTIVE SITE | | Descriptor: | MAGNESIUM ION, N-METHYLMESOPORPHYRIN CONTAINING COPPER, PROTOHEME FERROLYASE | | Authors: | Lecerof, D, Fodje, M.N, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
1C9I
 
 | |
