1RM5
 
 | | Crystal structure of mutant S188A of photosynthetic glyceraldehyde-3-phosphate dehydrogenase A4 isoform, complexed with NADP | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Sparla, F, Fermani, S, Falini, G, Ripamonti, A, Sabatino, P, Pupillo, P, Trost, P. | | Deposit date: | 2003-11-27 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Coenzyme Site-directed Mutants of Photosynthetic A(4)-GAPDH Show Selectively Reduced NADPH-dependent Catalysis, Similar to Regulatory AB-GAPDH Inhibited by Oxidized Thioredoxin
J.Mol.Biol., 340, 2004
|
|
1RM6
 
 | | Structure of 4-hydroxybenzoyl-CoA reductase from Thauera aromatica | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxybenzoyl-CoA reductase alpha subunit, ... | | Authors: | Unciuleac, M, Warkentin, E, Page, C.C, Dutton, P.L, Boll, M, Ermler, U. | | Deposit date: | 2003-11-27 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Xanthine Oxidase-Related 4-Hydroxybenzoyl-CoA Reductase with an Additional [4Fe-4S] Cluster and an Inverted Electron Flow
Structure, 12, 2004
|
|
1RM9
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | avermectin-sensitive chloride channel GluCl beta/cyan fluorescent protein fusion | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-27 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
1RMD
 
 | | RAG1 DIMERIZATION DOMAIN | | Descriptor: | RAG1, ZINC ION | | Authors: | Bellon, S.F, Rodgers, K.K, Schatz, D.G, Coleman, J.E, Steitz, T.A. | | Deposit date: | 1997-01-10 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster.
Nat.Struct.Biol., 4, 1997
|
|
1RMF
 
 | |
1RMG
 
 | | RHAMNOGALACTURONASE A FROM ASPERGILLUS ACULEATUS | | Descriptor: | RHAMNOGALACTURONASE A, alpha-D-glucopyranose, alpha-D-mannopyranose, ... | | Authors: | Petersen, T.N, Kauppinen, S, Larsen, S. | | Deposit date: | 1997-02-26 | | Release date: | 1998-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed parallel beta helix.
Structure, 5, 1997
|
|
1RMQ
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase with osmiate mimicking the catalytic intermediate | | Descriptor: | COBALT (II) ION, Class B acid phosphatase, OSMIUM ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights in the catalytic mechanism of AphA from Escherichia coli
To be Published
|
|
1RMT
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase complexed with adenosine. | | Descriptor: | ADENOSINE, Class B acid phosphatase, MAGNESIUM ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights in the catalytic mechanism of AphA from Escherichia coli
To be Published
|
|
1RMU
 
 | | THREE-DIMENSIONAL STRUCTURES OF DRUG-RESISTANT MUTANTS OF HUMAN RHINOVIRUS 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Krishnaswamy, S, Kremer, M.J, Oliveira, M.A, Rossmann, M.G, Heinz, B.A, Rueckert, R.R, Dutko, F.J, Mckinlay, M.A. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of drug-resistant mutants of human rhinovirus 14.
J.Mol.Biol., 207, 1989
|
|
1RMY
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase ternary complex with deoxycytosine and phosphate bound to the catalytic metal | | Descriptor: | 2'-DEOXYCYTIDINE, Class B acid phosphatase, MAGNESIUM ION, ... | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights in the catalytic mechanism of AphA from Escherichia coli
To be Published
|
|
1RMZ
 
 | | Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor NNGH at 1.3 A resolution | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Conformational variability of matrix metalloproteinases: beyond a single 3D structure.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1RN7
 
 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-11-30 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
1RN8
 
 | | Crystal structure of dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
1RNC
 
 | | NEWLY OBSERVED BINDING MODE IN PANCREATIC RIBONUCLEASE | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE A, SULFATE ION | | Authors: | Aguilar, C.F, Thomas, P.J, Mills, A, Moss, D.S, Palmer, R.A. | | Deposit date: | 1991-10-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Newly observed binding mode in pancreatic ribonuclease.
J.Mol.Biol., 224, 1992
|
|
1RND
 
 | | NEWLY OBSERVED BINDING MODE IN PANCREATIC RIBONUCLEASE | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE A, SULFATE ION | | Authors: | Aguilar, C.F, Thomas, P.J, Mills, A, Moss, D.S, Palmer, R.A. | | Deposit date: | 1991-10-21 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Newly observed binding mode in pancreatic ribonuclease.
J.Mol.Biol., 224, 1992
|
|
1RNI
 
 | | Bifunctional DNA primase/polymerase domain of ORF904 from the archaeal plasmid pRN1 | | Descriptor: | ORF904, ZINC ION | | Authors: | Lipps, G, Weinzierl, A.O, von Scheven, G, Buchen, C, Cramer, P. | | Deposit date: | 2003-12-01 | | Release date: | 2004-01-27 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a bifunctional DNA primase-polymerase
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RNJ
 
 | | Crystal structure of inactive mutant dUTPase complexed with substrate analogue imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
1RNL
 
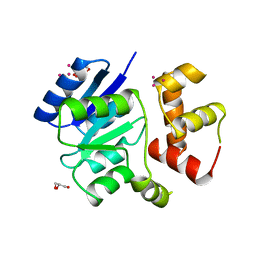 | | THE NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL FROM NARL | | Descriptor: | GLYCEROL, NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL, PLATINUM (II) ION | | Authors: | Baikalov, I, Schroder, I, Kaczor-Grzeskowiak, M, Grzeskowiak, K, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 1996-04-17 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Escherichia coli response regulator NarL.
Biochemistry, 35, 1996
|
|
1RNT
 
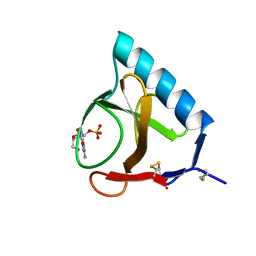 | | RESTRAINED LEAST-SQUARES REFINEMENT OF THE CRYSTAL STRUCTURE OF THE RIBONUCLEASE T1(ASTERISK)2(PRIME)-GUANYLIC ACID COMPLEX AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 ISOZYME | | Authors: | Saenger, W, Arni, R, Heinemann, U, Tokuoka, R. | | Deposit date: | 1987-07-10 | | Release date: | 1987-10-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restrained Least-Squares Refinement of the Crystal Structure of the Ribonuclease T1(Asterisk)2(Prime)-Guanylic Acid Complex at 1.9 Angstroms Resolution
Acta Crystallogr.,Sect.B, 43, 1987
|
|
1RO0
 
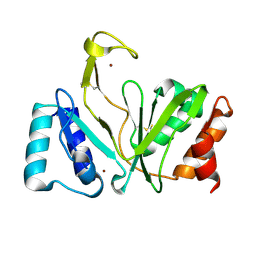 | | Bifunctional DNA primase/polymerase domain of ORF904 from the archaeal plasmid pRN1- Triple mutant F50M/L107M/L110M SeMet remote | | Descriptor: | ORF904, ZINC ION | | Authors: | Lipps, G, Weinzierl, A.O, von Scheven, G, Buchen, C, Cramer, P. | | Deposit date: | 2003-12-01 | | Release date: | 2004-01-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a bifunctional DNA primase-polymerase
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RO2
 
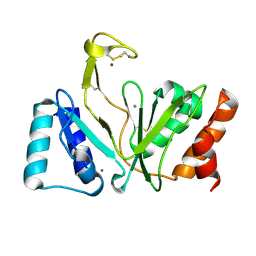 | | Bifunctional DNA primase/polymerase domain of ORF904 from the archaeal plasmid pRN1- Triple mutant F50M/L107M/L110M manganese soak | | Descriptor: | MANGANESE (II) ION, ZINC ION, hypothetical protein ORF904 | | Authors: | Lipps, G, Weinzierl, A.O, von Scheven, G, Buchen, C, Cramer, P. | | Deposit date: | 2003-12-01 | | Release date: | 2004-01-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a bifunctional DNA primase-polymerase
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RO6
 
 | | Crystal structure of PDE4B2B complexed with Rolipram (R & S) | | Descriptor: | ARSENIC, MANGANESE (II) ION, ROLIPRAM, ... | | Authors: | Xu, R.X, Rocque, W.J, Lambert, M.H, Vanderwall, D.E, Nolte, R.T. | | Deposit date: | 2003-12-01 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram.
J.Mol.Biol., 337, 2004
|
|
1RO7
 
 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, CMP-3FNeuAc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RO8
 
 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, cytidine-5'-monophosphate | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RO9
 
 | | CRYSTAL STRUCTURES OF THE CATALYTIC DOMAIN OF PHOSPHODIESTERASE 4B2B COMPLEXED WITH 8-Br-AMP | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4B | | Authors: | Xu, R.X, Rocque, W.J, Lambert, M.H, Vanderwall, D.E, Nolte, R.T. | | Deposit date: | 2003-12-01 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram.
J.Mol.Biol., 337, 2004
|
|
